Michal Neumayr,
Michael Stary,
Simon Pannek,
Daniel Korth
Technical University of Munich
- Diverse Views: Enhance Semantic Scene Understanding by selecting more diverse views for the CLIP Embeddings
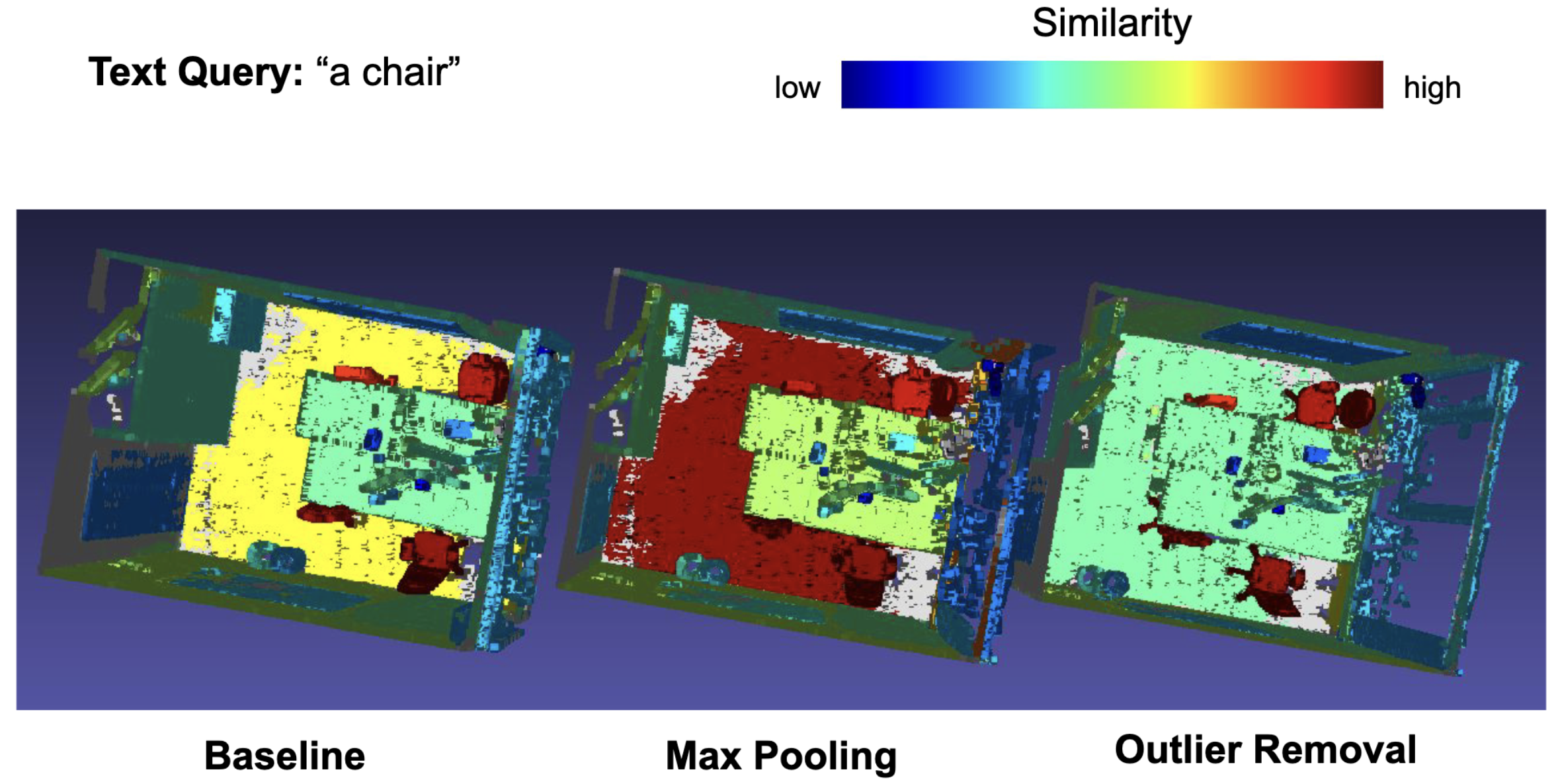
- Outliers Removal: removing outliers in the embedding space to get more consistent semantic signals and experimenting with different aggregation fucntions
- Foundation Models: Replace CLIP with SigLip and use DINOv2 to allow image queres of the 3D Scene
For setting up OpenMask3D, please refer to the official GitHub and follow their steps at from here.
Once you have OpenMask3D installed and running, you need to download ScanNet++ and preprocess each scene with the following command:
python scannet++_to_openmask.py --scene_id $scene_idMake sure to set the replace the paths in the file to match your local setup.
You can try out modifications by running the script with following arguments:
path_to_scenes_txt: the path to the scenes you want to have processednum_views_precomputed: 5 is the default from OpenMask3D. If you use more than 5, then it will select the 5 images per instance that will make it most diverse (in terms of camera angles)model: the foundation model to use. Select betweenclip,siglipanddinov2
bash run_openmask3d_selected_scenes_scannet++.sh $path_to_scenes_txt $num_views_precomputed $modelOnce you computed the instance masks for each scene, you can query it either via text or image. Querying via text works with clip and siglip, querying via image with dinov2
Following arguments need to be provide:
experiment_path: Path to the scene in the respective experiment folder.text: The text to query.remove_outliers: whether to remove outlier embeddings or not. Default is False.agg_fct: Aggregation function over the embeddings; choose betweenmean(default) andmax.
python openmask3d/visualization/vim_sim_score_export.py \
-e $experiment_path \
-t $text \
--remove_outliers $remove_outliers \
--agg_fct $agg_fctIf you want to query by image, replace -t $text with -i path_to_image.
To quantitatively evaluate the results, use the following script with additional an additional argument topk which indicates whether you want to evaluate the model on top1, top5, or other topk. We use 1 for ScanNet200 and 5 for ScanNet++.
bash run_evaluation_scannet++.sh $experiment_path $remove_outliers $agg_fct $topk