IEDB Protein Tree
Assigning IEDB source antigens and epitopes to their genes and proteins.
Current Success Rates:
- Source Antigen Assignment: 97.9%
- Epitope Assignment: 87.4%
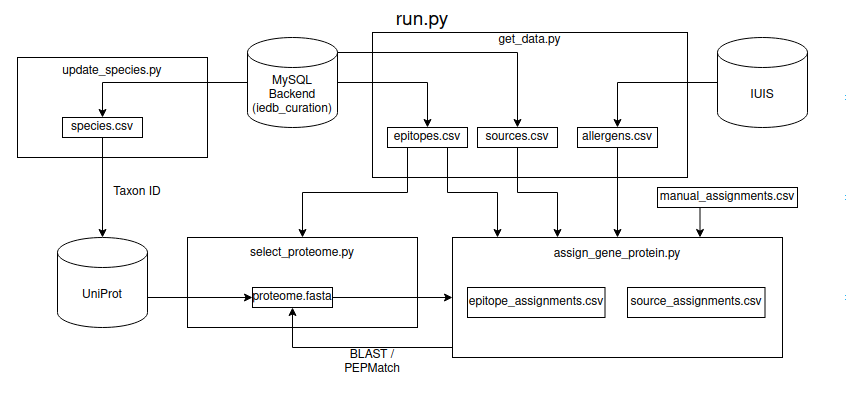
Process
- Collect the epitope and source antigen data for a species.
- Select the best proteome for that species from UniProt.
- Assign gene/protein to source antigens and epitopes using BLAST, ARC, and PEPMatch.
Inputs
- IEDB MySQL backend access
- List of IEDB species: species.csv
- This is updated with update_species.py
blastpandmakeblastdbbinaries from NCBIhmmscanbinary from HMMER- manual_assignments.csv - manually assigned proteins
allergens.csv- IUIS allergen nomenclature; get/update using get_data.py- Flags (for run.py)
-a- run for all species-t- run for a single species using its taxon ID-d- update epitope, source antigen, and allergen data-p- update proteome to be used for the species-s- update species list (runs update_species.py)-n- number of threads to speed up source antigen assignment
Running
To run the entire pipeline:
- for one species:
protein_tree/run.py -t <taxon ID>- for all species:
protein_tree/run.py -aGetting the raw epitope and source antigen data can be run separately:
protein_tree/get_data.py -t <taxon ID>Selecting the best proteome can also be run separately:
protein_tree/select_proteome.py -t <taxon ID>Outputs
For each species:
- proteome.fasta - selected proteome in FASTA
- source_assignments.csv - each source antigen with assigned gene and protein
- epitope_assignments.csv - each epitope with its source antigen and assigned protein
- [optional] gp_proteome.fasta - the gene priority proteome in FASTA if it exists
For all species:
- metrics.csv - the metadata from the build
- Proteome ID
- Proteome Taxon
- Proteome Type
- Source Antgigen Count
- Epitope Count
- Successful Source Assignement (%)
- Successful Epitope Assignment (%)
- all_epitope_assinments.csv - combined epitope assignments for every species
- all_source_assignments.csv - combined source antigen assignments for every species
Use combine_data.py to merge all the assignments into the all_epitope_assignments.csv and all_source_assignments.csv files.
TODO
- Create a tree for visualization