Code to accompany Edwards et al, 2023, https://doi.org/10.7554/eLife.83364
Edwards, C.A., Watkisnon, W.M.D., Telerman, S.B., Hulsmann, L.C., Hamilton, R.S. & Ferguson-Smith, A.C. (2023) Reassessment of weak parent-of-origin expression bias shows it rarely exists outside of known imprinted regions. eLife 12:e83364 [eLife] [DOI]
Department of Genetics, University of Cambridge, Downing Street, Cambridge, CB2 3EHbioRxiv preprint available at: https://doi.org/10.1101/2022.08.21.504536
Raw sequencing read FASTQ files were downloaded from EMBL-EBI European Nucleotide Archive for each of the RNA-seq data sets ( [DOI] [DOI] [DOI] [DOI] [DOI] ). Low quality bases and adapters were removed with trim_galore (v0.4.1) [WEB]. SNPSplit (v0.3.4) [DOI] was used to separate reads by parent of origin, which first required the preparation of allele-specific reference genomes for C57BL6/CAST_Eij and CAST_Eij/FVB (based on C57BL6) with the following commands SNPsplit_genome_preparation --vcf_file mgp.v5.merged.snps_all.dbSNP142.vcf.gz --reference_genome GRCm38_fasta/ --strain CAST_EiJ and SNPsplit_genome_preparation --vcf_file mgp.v5.merged.snps_all.dbSNP142.vcf.gz --reference_genome GRCm38_fasta/ --strain CAST_EiJ --strain2 FVB_NJ --dual_hybrid. VCF files for strain specific SNPs were obtained from www.sanger.ac.uk/data/mouse-genomes-project. The Clusterflow pipeline tool was used to enable running multiple jobs in parallel across multiple processors on a HPC, however all scripts are also compatible with running on a single processor [DOI]. Trimmed reads were aligned to either the C57BL6/CAST_Eij or CAST_Eij/FVB reference genomes using HiSat2 (v2.1.0)[DOI], run via the hisat2 ClusterFlow module. Aligned reads were then name sorted to be compatible with SNPSplit, run via the samtools (v1.9)[DOI] Clusterflow module. Aligned files were run through SNPSplit to produce separated parent-specific alignment files using the SNP files produced by the genome preparation: all_SNPs_CAST_EiJ_GRCm38.txt.gz or all_FVB_NJ_SNPs_CAST_EiJ_reference.based_on_GRCm38.txt.gz. A custom Clusterflow module was created for SNPsplit [SNPSplit.cfmod]. Gene counts from each parent-specific alignment BAM file produced by SNPSplit were calculated using featureCounts (v1.5.0-p2)[DOI] via Clusterflow. A custom Rscript DESeq2_featureCounts_2_CountsTables.R is used to make a single counts table, including normalised reads, from the individual featureCount files. All scripts to reproduce the analysis are freely available at github.com/darogan/ASE_Meta_Analysis.
A sanity check to make sure all the samples behave in a similar, or expected manner
| Dataset | Read Counts |
|---|---|
| Andergassen_2015 |  [PDF] |
| Babak_2015 |  [PDF] |
| Bonthuis_2015 DRN TO ADD |
 [PDF] |
| Perez_2015 |  [PDF] |
| Teichman_ESCs_NPCs |  [PDF] |
| Dataset | Publication | Details |
|---|---|---|
| Andergassen_2015 | [DOI] | Cast/EiJ X FVB strains |
| Babak_2015 | [DOI] | C57Bl/6J x Cast (maternal allele first) |
| Bonthuis_2015 | [DOI] | C57Bl/6J x Cast (maternal allele first) |
| Perez_2015 | [DOI] | F1i (F1 hybrid) C57Bl/6J father and Cast/EiJ mother = CB ; F1r (F1 hybrid) Cast/EiJ father and C57Bl/6J mother = BC |
| Teichman_ESCs_NPCs | [DOI] | Cell: C57Bl/6J and Cast/EiJ (maternal allele first) |
Andergassen_2015
| Accession | Tissue | Cross | Replicate |
|---|---|---|---|
| SRR3085966 | adult Brain | CF | 1 |
| SRR3085967 | adult Brain | CF | 2 |
| SRR3085968 | adult Brain | FC | 1 |
| SRR3085969 | adult Brain | FC | 2 |
| SRR3085970 | adult Liver | CF | 1 |
| SRR3085971 | adult Liver | CF | 2 |
| SRR3085972 | adult Liver | FC | 1 |
| SRR3085973 | adult Liver | FC | 2 |
| SRR3085990 | adult Leg Muscle | CF | 1 |
| SRR3085991 | adult Leg Muscle | CF | 2 |
| SRR3085992 | adult Leg Muscle | FC | 1 |
| SRR3085993 | adult Leg Muscle | FC | 2 |
Babak_2015
| Accession | Tissue | Cross | Replicate |
|---|---|---|---|
| SRR823449 | Muscle | CB | 1 |
| SRR823450 | Muscle | BC | 1 |
| SRR823469 | Liver | BC | 1 |
| SRR823474 | Liver | CB | 1 |
| SRR823485 | Hypothalamus | CB | 1 |
| SRR823478 | Hypothalamus | BC | 1 |
| SRR823461 | Cerebellum | BC | 1 |
| SRR823458 | Cerebellum | CB | 1 |
| SRR823472 | Adult Whole Brain | CB | 1 |
| SRR823473 | Adult Whole Brain | BC | 1 |
Bonthuis_2015
| Accession | Tissue | Cross | Replicate |
|---|---|---|---|
| SRR2086215 | ARN | BC | 1 |
| SRR2086216 | ARN | BC | 2 |
| SRR2086217 | ARN | BC | 3 |
| SRR2086218 | ARN | BC | 4 |
| SRR2086219 | ARN | BC | 5 |
| SRR2086220 | ARN | BC | 6 |
| SRR2086221 | ARN | BC | 7 |
| SRR2086222 | ARN | BC | 8 |
| SRR2086223 | ARN | BC | 9 |
| SRR2086224 | ARN | CB | 1 |
| SRR2086225 | ARN | CB | 2 |
| SRR2086226 | ARN | CB | 3 |
| SRR2086227 | ARN | CB | 4 |
| SRR2086228 | ARN | CB | 5 |
| SRR2086229 | ARN | CB | 6 |
| SRR2086230 | ARN | CB | 7 |
| SRR2086231 | ARN | CB | 8 |
| SRR2086232 | ARN | CB | 9 |
| SRR2086233 | DRN | BC | 1 |
| SRR2086234 | DRN | BC | 2 |
| SRR2086235 | DRN | BC | 3 |
| SRR2086236 | DRN | BC | 4 |
| SRR2086237 | DRN | BC | 5 |
| SRR2086238 | DRN | BC | 6 |
| SRR2086239 | DRN | BC | 7 |
| SRR2086240 | DRN | BC | 8 |
| SRR2086241 | DRN | BC | 9 |
| SRR2086242 | DRN | CB | 1 |
| SRR2086243 | DRN | CB | 2 |
| SRR2086244 | DRN | CB | 3 |
| SRR2086245 | DRN | CB | 4 |
| SRR2086246 | DRN | CB | 5 |
| SRR2086247 | DRN | CB | 6 |
| SRR2086248 | DRN | CB | 7 |
| SRR2086249 | DRN | CB | 8 |
| SRR2086250 | DRN | CB | 9 |
| SRR2086251 | Liver | BC | 1 |
| SRR2086252 | Liver | BC | 2 |
| SRR2086253 | Liver | BC | 3 |
| SRR2086254 | Liver | BC | 4 |
| SRR2086255 | Liver | BC | 5 |
| SRR2086256 | Liver | BC | 6 |
| SRR2086257 | Liver | BC | 7 |
| SRR2086258 | Liver | BC | 8 |
| SRR2086259 | Liver | CB | 1 |
| SRR2086260 | Liver | CB | 2 |
| SRR2086261 | Liver | CB | 3 |
| SRR2086262 | Liver | CB | 4 |
| SRR2086263 | Liver | CB | 5 |
| SRR2086264 | Liver | CB | 6 |
| SRR2086265 | Liver | CB | 7 |
| SRR2086266 | Liver | CB | 8 |
| SRR2086267 | Muscle | BC | 1 |
| SRR2086268 | Muscle | BC | 2 |
| SRR2086269 | Muscle | BC | 3 |
| SRR2086270 | Muscle | BC | 4 |
| SRR2086271 | Muscle | BC | 5 |
| SRR2086272 | Muscle | BC | 6 |
| SRR2086273 | Muscle | BC | 7 |
| SRR2086274 | Muscle | BC | 8 |
| SRR2086275 | Muscle | CB | 1 |
| SRR2086276 | Muscle | CB | 2 |
| SRR2086277 | Muscle | CB | 3 |
| SRR2086278 | Muscle | CB | 4 |
| SRR2086279 | Muscle | CB | 5 |
| SRR2086280 | Muscle | CB | 6 |
| SRR2086281 | Muscle | CB | 7 |
| SRR2086282 | Muscle | CB | 8 |
Perez_2015
| Accession | Tissue | Cross | Replicate |
|---|---|---|---|
| SRR1952382 | P8 Female | CB | 1 |
| SRR1952383 | P8 Female | CB | 2 |
| SRR1952384 | P8 Female | CB | 3 |
| SRR1952385 | P8 Female | CB | 4 |
| SRR1952386 | P8 Female | CB | 5 |
| SRR1952387 | P8 Female | CB | 6 |
| SRR1952388 | P8 Male | CB | 1 |
| SRR1952389 | P8 Male | CB | 2 |
| SRR1952390 | P8 Male | CB | 3 |
| SRR1952391 | P8 Male | CB | 4 |
| SRR1952392 | P8 Male | CB | 5 |
| SRR1952393 | P8 Male | CB | 6 |
| SRR1952394 | P8 Female | BC | 1 |
| SRR1952395 | P8 Female | BC | 2 |
| SRR1952396 | P8 Female | BC | 3 |
| SRR1952397 | P8 Female | BC | 4 |
| SRR1952398 | P8 Female | BC | 5 |
| SRR1952399 | P8 Female | BC | 6 |
| SRR1952400 | P8 Male | BC | 1 |
| SRR1952401 | P8 Male | BC | 2 |
| SRR1952402 | P8 Male | BC | 3 |
| SRR1952403 | P8 Male | BC | 4 |
| SRR1952404 | P8 Male | BC | 5 |
| SRR1952405 | P8 Male | BC | 6 |
| SRR1952406 | P60 Female | CB | 1 |
| SRR1952407 | P60 Female | CB | 2 |
| SRR1952408 | P60 Female | CB | 3 |
| SRR1952409 | P60 Female | CB | 4 |
| SRR1952410 | P60 Female | CB | 5 |
| SRR1952411 | P60 Female | CB | 6 |
| SRR1952412 | P60 Male | CB | 1 |
| SRR1952413 | P60 Male | CB | 2 |
| SRR1952414 | P60 Male | CB | 3 |
| SRR1952415 | P60 Male | CB | 4 |
| SRR1952416 | P60 Male | CB | 5 |
| SRR1952417 | P60 Male | CB | 6 |
| SRR1952419 | P60 Female | BC | 1 |
| SRR1952420 | P60 Female | BC | 2 |
| SRR1952421 | P60 Female | BC | 3 |
| SRR1952422 | P60 Female | BC | 4 |
| SRR1952423 | P60 Female | BC | 5 |
| SRR1952424 | P60 Female | BC | 6 |
| SRR1952425 | P60 Male | BC | 1 |
| SRR1952426 | P60 Male | BC | 2 |
| SRR1952427 | P60 Male | BC | 3 |
| SRR1952428 | P60 Male | BC | 4 |
| SRR1952429 | P60 Male | BC | 5 |
| SRR1952430 | P60 Male | BC | 6 |
Teichman_ESCs_NPCs
| Accession | Tissue | Cross | Replicate |
|---|---|---|---|
| SRR6330118 | ESCs | BC8 | |
| SRR6330119 | ESCs | CB9 | |
| SRR6330120 | neural precursor cell (day 3) | BC8 | |
| SRR6330121 | neural precursor cell (day 3) | CB9 | |
| SRR6330122 | neural precursor cell (day 6) | BC8 | |
| SRR6330123 | neural precursor cell (day 6) | CB9 | |
| SRR6330124 | neural precursor cell (day 8) | BC8 | |
| SRR6330125 | neural precursor cell (day 8) | CB9 |
| Dataset | WGET commands |
|---|---|
| Andergassen_2015 | Andergassen_2015.wget.sh |
| Babak_2015 | Babak_2015.wget.sh |
| Bonthuis_2015 | Bonthuis_2015.wget.sh |
| Perez_2015 | Perez_2015.wget.sh |
| Teichman_ESCs_NPCs | Teichman_ESCs_NPCs.wget.sh |
Using TrimGalore! run via the ClusterFlow pipeline tool. Single and paired-end reads are automatically determined and run accordingly.
cf trim_galore *.fq.gz or cf trim_galore *.fastq.gz
[mgp.v5.merged.snps_all.dbSNP142.vcf.gz]
Assumes a directory, GRCm38_fasta, containing the BL6 GRCm38 reference genome fasta files
BL6 Vs CAST
SNPsplit_genome_preparation --vcf_file mgp.v5.merged.snps_all.dbSNP142.vcf.gz --reference_genome GRCm38_fasta/ --strain CAST_EiJ
CAST Vs FVB (based on GRCm38)
SNPsplit_genome_preparation --vcf_file mgp.v5.merged.snps_all.dbSNP142.vcf.gz --reference_genome GRCm38_fasta/ --strain CAST_EiJ --strain2 FVB_NJ --dual_hybrid
cf --genome CAST_EiJ_N-masked hisat2 *.fq.gz
cf --genome CAST_EiJ_FVB_NJ_dual_hybrid.based_on_GRCm38_N-masked hisat2 *.fq.gz
cf --params byname samtools_sort *N-masked_hisat2.bam
Run via a custom clusterflow module [SNPSplit.cfmod.pl]](Code/SNPSplit.cfmod.pl), or command line.
For single-end reads: BL6 Vs CAST
cf --genome all_SNPs_CAST_EiJ_GRCm38 --params sorted,paired SNPSplit *N-masked_hisat2_srtd.bam
For single-end reads: CAST Vs FVB
cf --genome all_FVB_NJ_SNPs_CAST_EiJ_reference.based_on_GRCm38 --params sorted SNPSplit *N-masked_hisat2_srtd.bam
For Paired-end reads: BL6 Vs CAST
cf --genome all_SNPs_CAST_EiJ_GRCm38 --params sorted,paired SNPSplit *N-masked_hisat2_srtd.bam
For Paired-end reads: CAST Vs FVB
cf --genome all_FVB_NJ_SNPs_CAST_EiJ_reference.based_on_GRCm38 --params sorted,paired SNPSplit *N-masked_hisat2_srtd.bam
For BL6 Vs CAST crosses
SNPFILE="all_SNPs_CAST_EiJ_GRCm38.txt.gz"
For CAST Vs FVB crosses
SNPFILE="all_FVB_NJ_SNPs_CAST_EiJ_reference.based_on_GRCm38.txt.gz"
Run for all alignment files in a directory
for i in *N-masked_hisat2_srtd.bam;
do
SNPsplit --paired --no_sort --snp_file ${SNPFILE} ${i} &> ${i/.bam/.snpsplit.log}
done
Gene counts from alignment files are calculated using featureCounts
cf --genome GRCm38 featureCounts *genome[12].bam
A custom Rscript DESeq2_featureCounts_2_CountsTables.R is used to make a single counts table from the individual featureCount files.
Replace FOLDERNAME with the directory name containing the featureCount files.
Rscript DESeq2_featureCounts_2_CountsTables.R FOLDERNAME
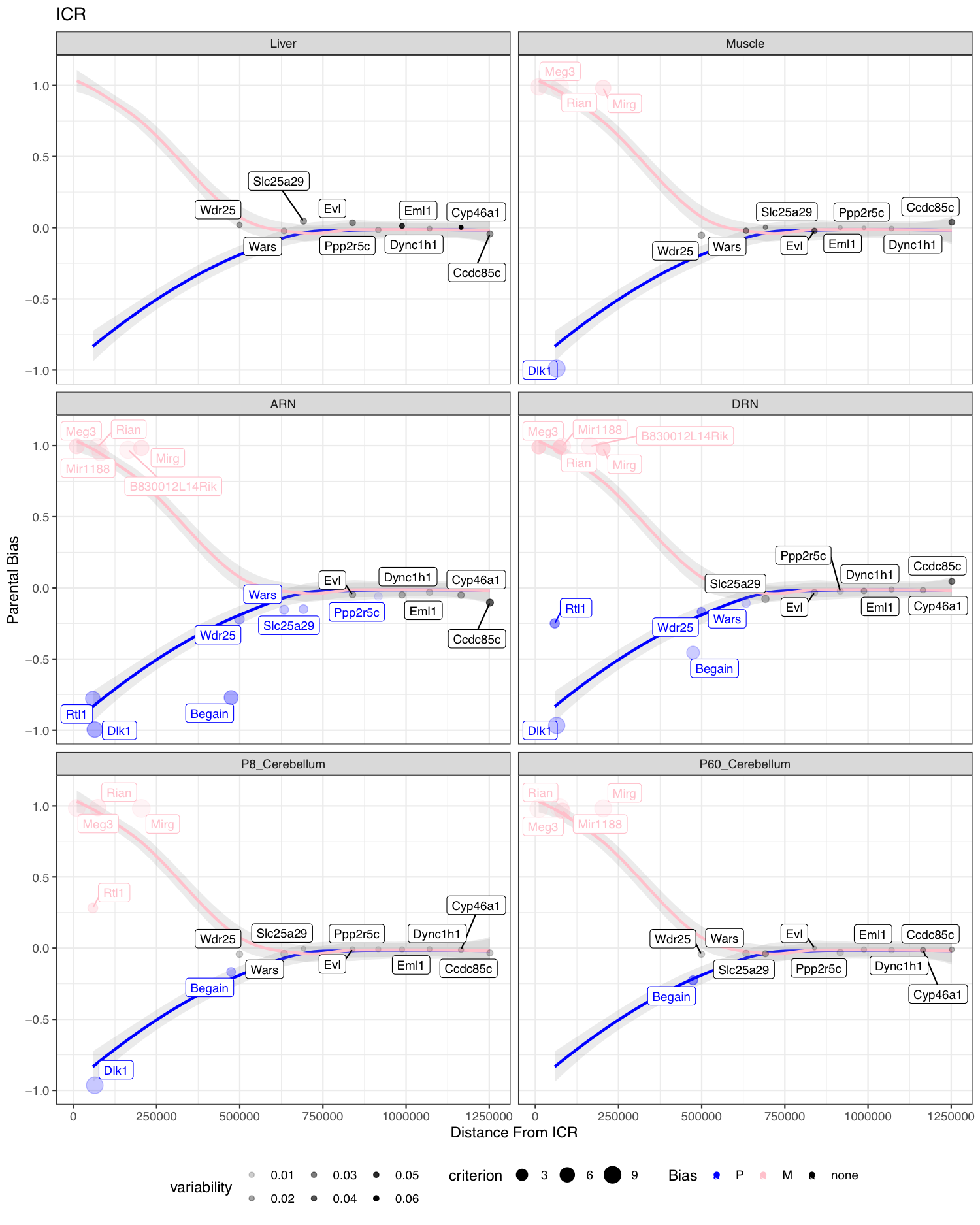
R code available in [ExploreBiasVsDist.R]. This is exploratory code and needs tidying up before publishing. Code works, but not commented or structured in a sensible way.
Needs completing
| Software | Version | Citation |
|---|---|---|
| ClusterFlow | v0.5 dev | [DOI] |
| TrimGalore! | v0.4.1 | |
| HISAT2 | v2.1.0 | [DOI] |
| samtools | v1.9 | [DOI] |
| featureCounts (subread) | v1.5.0-p2 | [DOI] |
| SNPsplit | v0.3.4 | [DOI |
| Description | URL |
|---|---|
| Preprint | https://doi.org/10.1101/2022.08.21.504536 |
| Publication | https://doi.org/10.7554/eLife.83364 |
Contact Russell S. Hamilton (darogan@gmail.com)