Continuous human uterine NK cell differentiation in response to endometrial regeneration and pregnancy
Benedikt Strunz1,‡, Jonna Bister1, Hanna Jönsson1, Iva Filipovic1, Ylva Crona-Guterstam1,2,3, Egle Kvedaraite1,4, Natalie Sleiers1, Bogdan Dumitrescu5, Mats Brännström6, Antonio Lentini7, Björn Reinius7, Martin Cornillet1, Tim Willinger1, Sebastian Gidlöf3,8, Russell S. Hamilton9,10, Martin A. Ivarsson1 & Niklas K. Björkström1,‡
1 Center for Infectious Medicine, Department of Medicine Huddinge, Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden
2Department of Clinical Science, Intervention and Technology, Karolinska Institutet, Stockholm, Sweden
3 Department of Obstetrics and Gynecology, Karolinska University Hospital Huddinge, Stockholm, Sweden
4 Childhood Cancer Research Unit, Department of Women’s and Children’s Health, Karolinska Institutet, Stockholm, Sweden
5 Department of Obstetrics and Gynecology, Mälarsjukhuset, Eskilstuna, Sweden
6 Department of Obstetrics and Gynecology, University of Gothenburg, Gothenburg, Swedenn
7 Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden
8 Department of Obstetrics and Gynecology, Stockholm South General Hospital, Stockholm, Sweden
9 Centre for Trophoblast Research, University of Cambridge, Cambridge, UK
10 Department of Genetics, University of Cambridge, Cambridge, UK
‡ Corresponding authors: benedikt.strunz@ki.se, niklas.bjorkstrom@ki.se
Strunz, B., Bister, J., Jönsson, H., Filipovic, I., Crona-Guterstam, Y., Kvedaraite, E., Sleiers, N., Dumitrescu, B., Brännström, M., Lentini, A., Reinius, B., Cornillet, M., Willinger, T., Gidlöf, S., Hamilton, R.S., Ivarsson, M.A., & Björkström, N.K. (2021) Continuous human uterine NK cell differentiation in response to endometrial regeneration and pregnancy. Science Immunology, 6, eabb7800 [Science Immunology] [DOI]
On publication
RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI under accession number E-MTAB-8709
| Sample Name | Group |
|---|---|
| 1_HU12_P11 | CD39+KIR+ |
| 2_HU12_P9 | CD39-KIR- |
| 3_HU12_P10 | CD39-KIR+ |
| 5_HU13_P11 | CD39+KIR+ |
| 6_HU13_P9 | CD39-KIR- |
| 7_HU13_P10 | CD39-KIR+ |
| 9_Hu05_P11 | CD39+KIR+ |
| 10_Hu05_P9 | CD39-KIR- |
| 11_Hu05_P10 | CD39-KIR+ |
| 14_Hu7_P12 | CD39+KIR+ |
| 15_Hu7_P13 | CD39-KIR+ |
| 16_Hu7_P9 | CD39-KIR- |
Raw sequencing files are run through quality control using FastQC (v0.11.5) and fastq_screen (v0.9.3). Low quality and adapter sequencing are trimmed with Trim Galore! (v0.6.4). Trimmed reads are aligned to the reference genome (GRCh38, ensEMBL) using STAR (v020201). Alignments are assessed using qualimap (v2.2) and featureCounts (v 1.5.0-p2). Gene quantification is performed with featureCounts (v 1.5.0-p2). Differential gene expression is performed with DESeq2 package (v1.22.2, R v3.5.3), including principle component analysis (PCA) to assess sample clustering, and multiple testing correction to produce false discovery rates. Finally all metrics from the RNA-Seq pipelines are summarised and reports produced using MultiQC (0.9.dev0).
Additional Processing Steps
Additional filtering steps were applied in order to focus on the high confidence DEGs. First, genes with exceeding inter-donor variation were excluded if the calculated CV2 (Anders & Huber 2010) surpassed a cut-off of CV2 >1.95 in the subset with highest read count. Next, genes with a mean expression of <10 reads were excluded and a FDR adjusted p-value <0.05 was used for determining significant DEGs.
Resources Used
| Resource | URL |
|---|---|
| GRCh38 | Link |
| FastQC | Link |
| Trim_galore | Link |
| STAR | DOI |
| HTSeq-counts | DOI |
| Feature_counts | DOI |
| Qualimap | DOI |
| RSeQC | DOI |
| ClusterFlow | DOI |
| MultiQC | DOI |
Pipeline run using ClusterFlow
#fastqc
#fastq_screen
#trim_galore
#fastqc
#star
#qualimap_rnaseq
#rseqc_infer_experiment
#featureCounts
#htseq_counts
FeatureCount gene count files are available in the FeatureCount directory
The R script used to generate the figures in the paper is RSH_KI_0001.QC.R available to download. Required R-packages are listed at the top of the file and must be installed prior to running the script.
| Figure | File | Description |
|---|---|---|
| Figure 2 F | 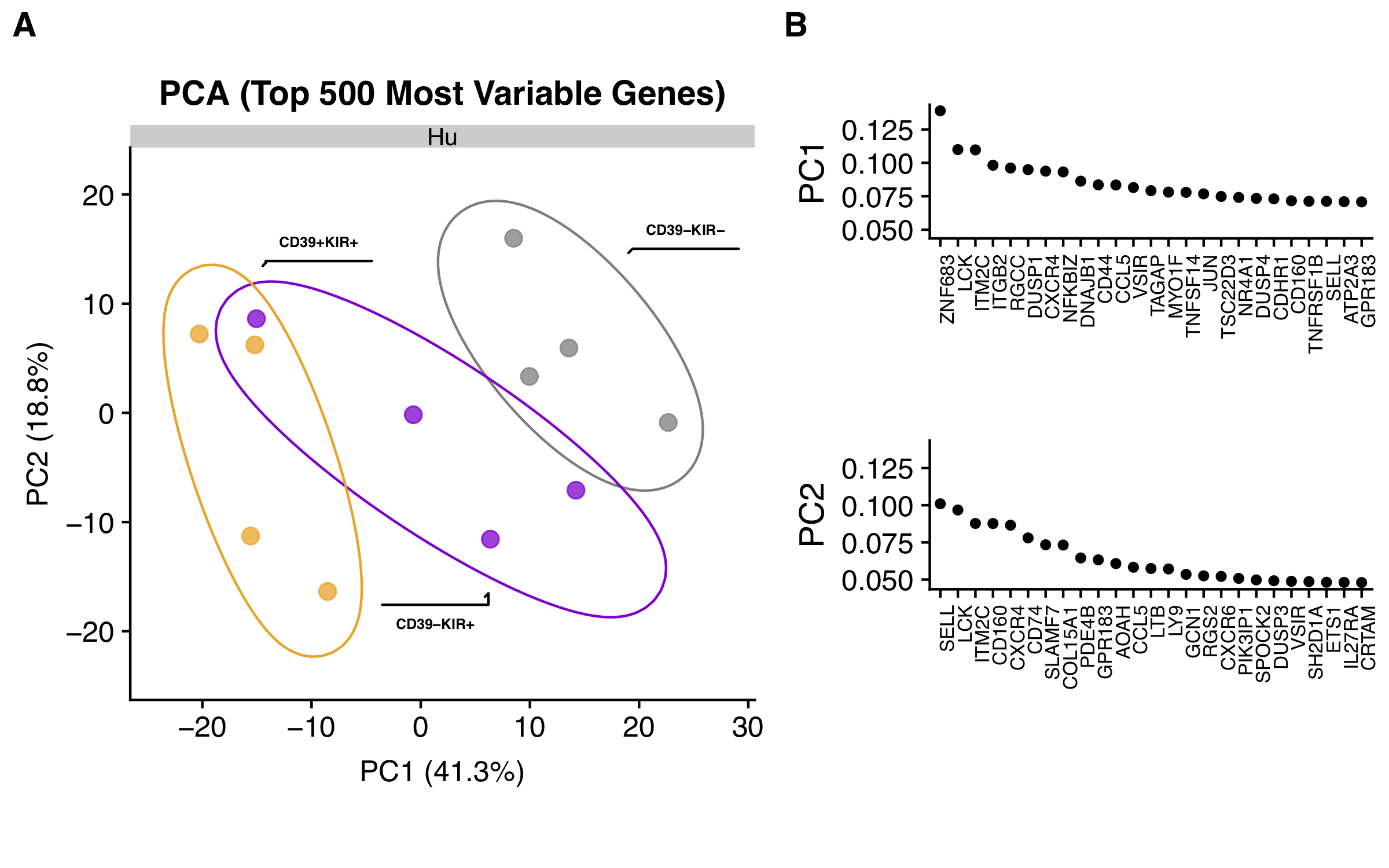 [PDF] [PNG] |
A. PCA for top 500 most variable genes |
| Figure 2 G (left) | 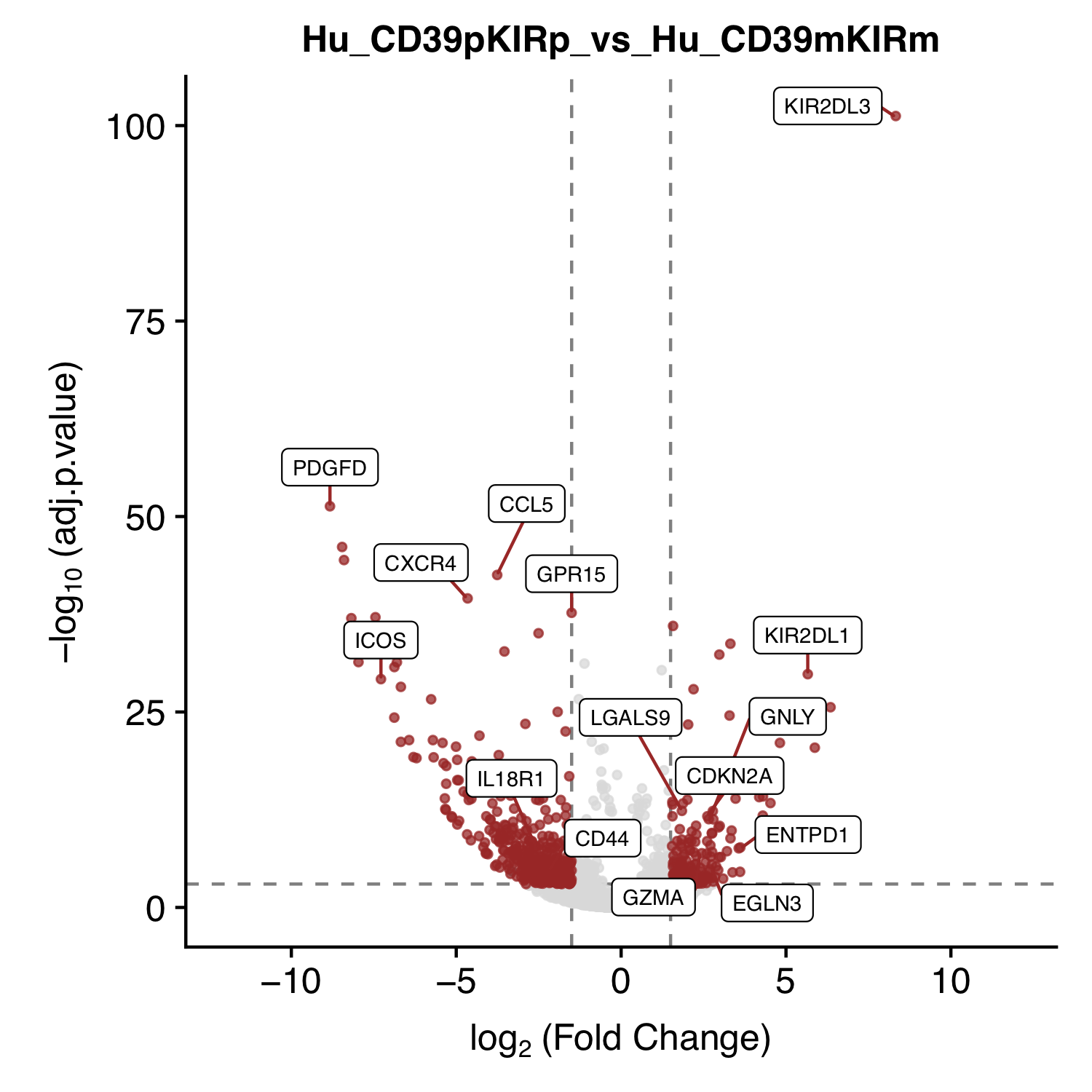 [PDF] [PNG] |
Volcano plot for KIR+CD39+ Vs KIR-CD39- |
| Figure 2 G (right) | 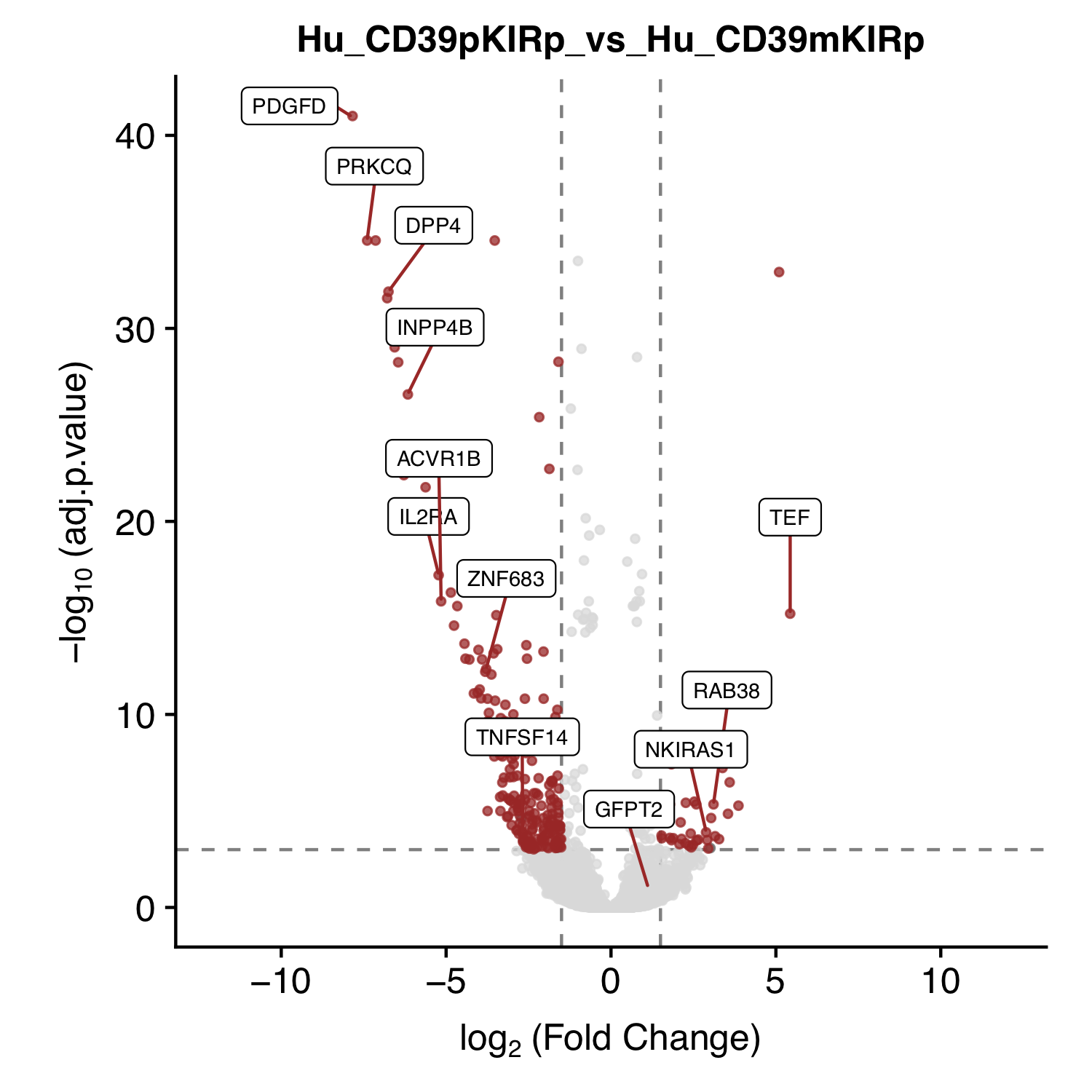 [PDF] [PNG] |
Volcano plot for KIR+CD39+ Vs KIR+CD39- |
| Figure S2 A |  [PDF] [PNG] |
B. PCA Principle components explained |
| Figure S2 B | 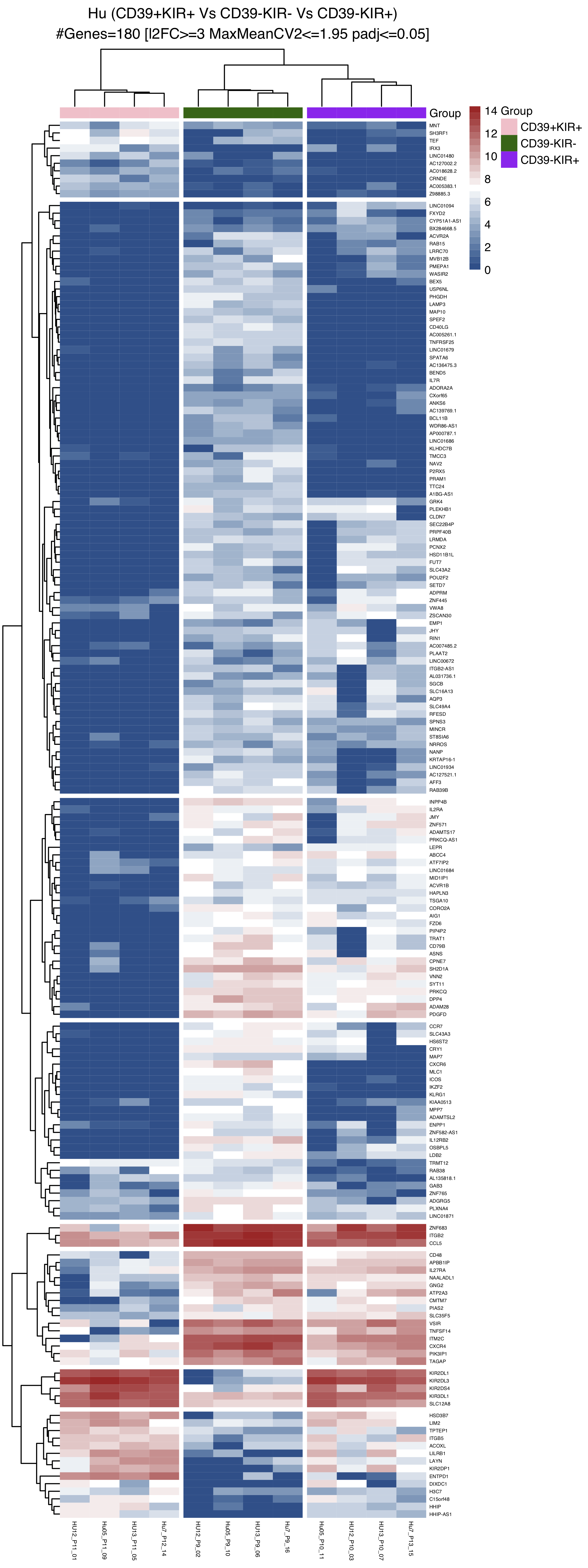 [PDF] [PNG] |
Heatmap summarising differentially expressed genes from the three comparisons |
| Figure S2 C | 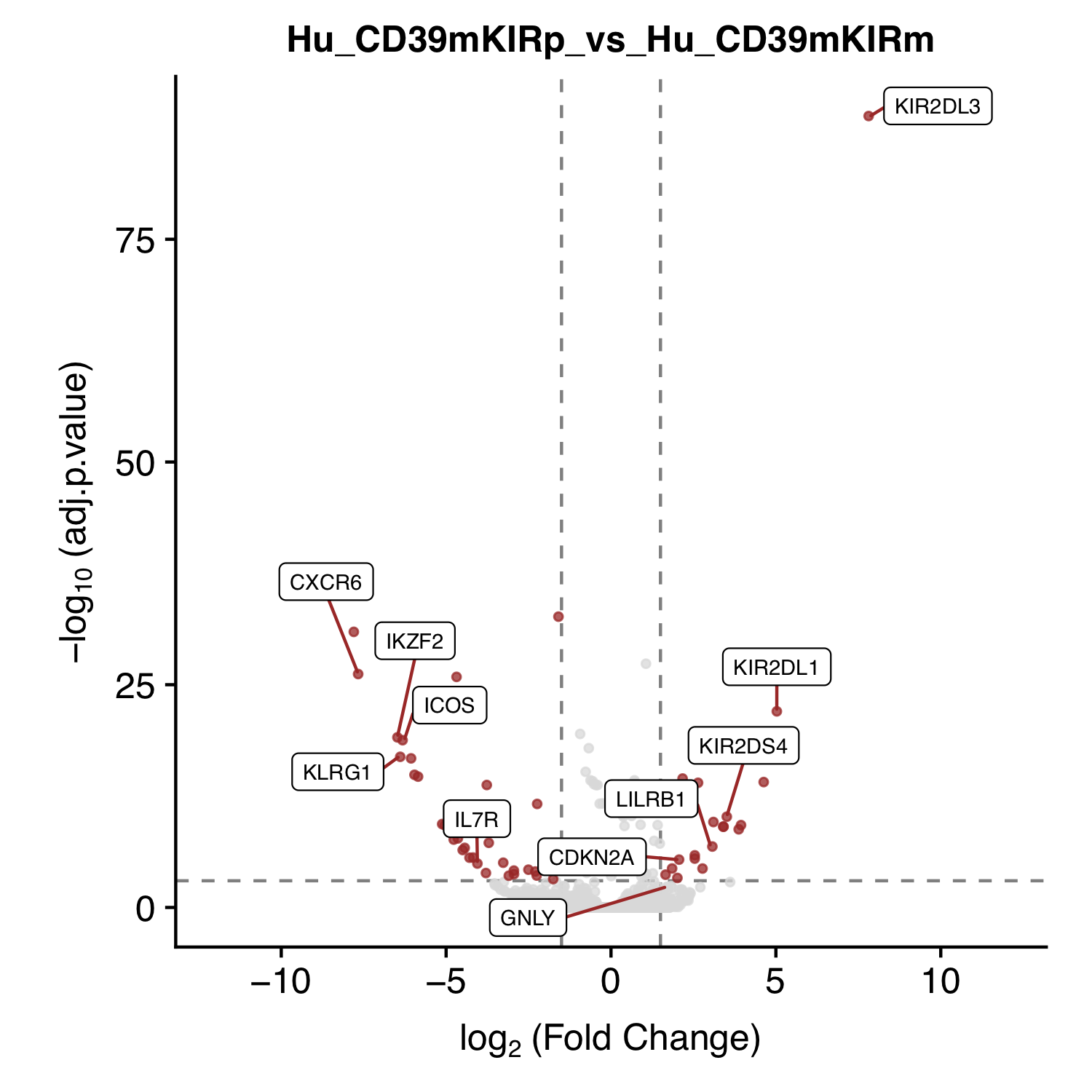 [PDF] [PNG] |
Volcano plot for KIR+CD39- Vs KIR-CD39- |
Normalised read counts [CSV]
| Comparison | Raw DEGs | CV2 Filtered | CV2 and MeanMax Filtered |
|---|---|---|---|
| Hu_CD39mKIRp_vs_Hu_CD39mKIRm | [CSV] | [CSV] | [CSV] |
| Hu_CD39pKIRp_vs_Hu_CD39mKIRm | [CSV] | [CSV] | [CSV] |
| Hu_CD39pKIRp_vs_Hu_CD39mKIRp | [CSV] | [CSV] | [CSV] |
Details for the R version and packages used to create all figures
> sessionInfo()
R version 3.4.4 (2018-03-15)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14.6
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] eulerr_5.1.0 RColorBrewer_1.1-2 pheatmap_1.0.12 Cairo_1.5-10 ggforce_0.3.1
[6] biomaRt_2.34.2 DESeq2_1.18.1 SummarizedExperiment_1.8.1 DelayedArray_0.4.1 Biobase_2.38.0
[11] GenomicRanges_1.30.3 GenomeInfoDb_1.14.0 IRanges_2.12.0 S4Vectors_0.16.0 BiocGenerics_0.24.0
[16] reshape2_1.4.3 reshape_0.8.8 useful_1.2.6 matrixStats_0.55.0 Matrix_1.2-17
[21] cowplot_0.9.4 ggrepel_0.8.1 ggplot2_3.2.1 dplyr_0.8.3 tidyr_1.0.0
loaded via a namespace (and not attached):
[1] bitops_1.0-6 bit64_0.9-7 progress_1.2.2 httr_1.4.1 tools_3.4.4 backports_1.1.4
[7] R6_2.4.0 rpart_4.1-15 Hmisc_4.2-0 DBI_1.0.0 lazyeval_0.2.2 colorspace_1.4-1
[13] nnet_7.3-12 withr_2.1.2 tidyselect_0.2.5 gridExtra_2.3 prettyunits_1.0.2 curl_4.2
[19] bit_1.1-14 compiler_3.4.4 htmlTable_1.13.2 labeling_0.3 scales_1.0.0 checkmate_1.9.4
[25] genefilter_1.60.0 stringr_1.4.0 digest_0.6.21 foreign_0.8-72 XVector_0.18.0 base64enc_0.1-3
[31] pkgconfig_2.0.3 htmltools_0.3.6 htmlwidgets_1.3 rlang_0.4.0 rstudioapi_0.10 RSQLite_2.1.2
[37] farver_1.1.0 BiocParallel_1.12.0 acepack_1.4.1 RCurl_1.95-4.12 magrittr_1.5 GenomeInfoDbData_1.0.0
[43] Formula_1.2-3 Rcpp_1.0.2 munsell_0.5.0 lifecycle_0.1.0 stringi_1.4.3 yaml_2.2.0
[49] MASS_7.3-51.4 zlibbioc_1.24.0 plyr_1.8.4 grid_3.4.4 blob_1.2.0 crayon_1.3.4
[55] lattice_0.20-38 splines_3.4.4 annotate_1.56.2 hms_0.5.1 locfit_1.5-9.1 zeallot_0.1.0
[61] knitr_1.25 pillar_1.4.2 geneplotter_1.56.0 XML_3.98-1.20 glue_1.3.1 latticeExtra_0.6-28
[67] data.table_1.11.8 vctrs_0.2.0 tweenr_1.0.1 gtable_0.3.0 purrr_0.3.2 polyclip_1.10-0
[73] assertthat_0.2.1 xfun_0.9 xtable_1.8-4 survival_2.44-1.1 tibble_2.1.3 AnnotationDbi_1.40.0
[79] memoise_1.1.0 cluster_2.1.0
| Description | URL |
|---|---|
| Publication | [Science Immunology] [DOI] |
| Raw Data | ArrayExpress EMBL-EBI E-MTAB-8709 |
| Björkström Group | Björkström group website |
| CTR Bioinformatics | CTR-BFX |
Contact Russell S. Hamilton (rsh46 -at- cam.ac.uk) for bioinformatics related queries