AbSeq is a comprehensive bioinformatic pipeline for the analysis of sequencing datasets generated from antibody libraries and abseqR is one of its packages.
abseqR empowers the users of abseqPy with
plotting and reporting capabilities and
allows them to generate interactive HTML reports for the convenience of viewing and sharing with other researchers. Additionally, abseqR extends abseqPy to compare multiple
repertoire analyses and perform further downstream analysis on its output.
Note that abseqR does not require abseqPy to be installed to work yet the output of abseqPy must be already present to invoke abseqR's functions.
-
AbSeq is developed by JiaHong Fong and Monther Alhamdoosh
-
For comments and suggestions, email jiahfong <at> gmail <dot> com
pandoc is highly
recommended. If abseqR fails to detect pandoc in your
system's PATH variable, it will not be able to generate a collated HTML report yet you can still access individual plots.
Make sure
pandocis at least version 1.19.2.1
As soon as abseqR is available in the Bioconductor project, it can be installed from R console as follows:
if (!require("BiocManager"))
install.packages("BiocManager")
BiocManager::install("abseqR")It is also possible to directly install abseqR from GitHub via devtools by running the following commands in R Console:
if (!require("devtools"))
install.packages("devtools")
devtools::install_github("malhamdoosh/abseqR")Assuming abseqPy has completed an analysis and the output directory was
named output, a basic usage of abseqR is:
library(abseqR)
abseqReport("/path/to/output")This will generate an interctive report of the analysis results. Please refer to the package's main vignette for more advanced use cases.
abseqR generates numerous plots for the quality control analysis of antibody libraries
and collates the results into an interactive HTML report. Here are some examples:
| V-(D)-J germline abundance | Diversity estimation |
|---|---|
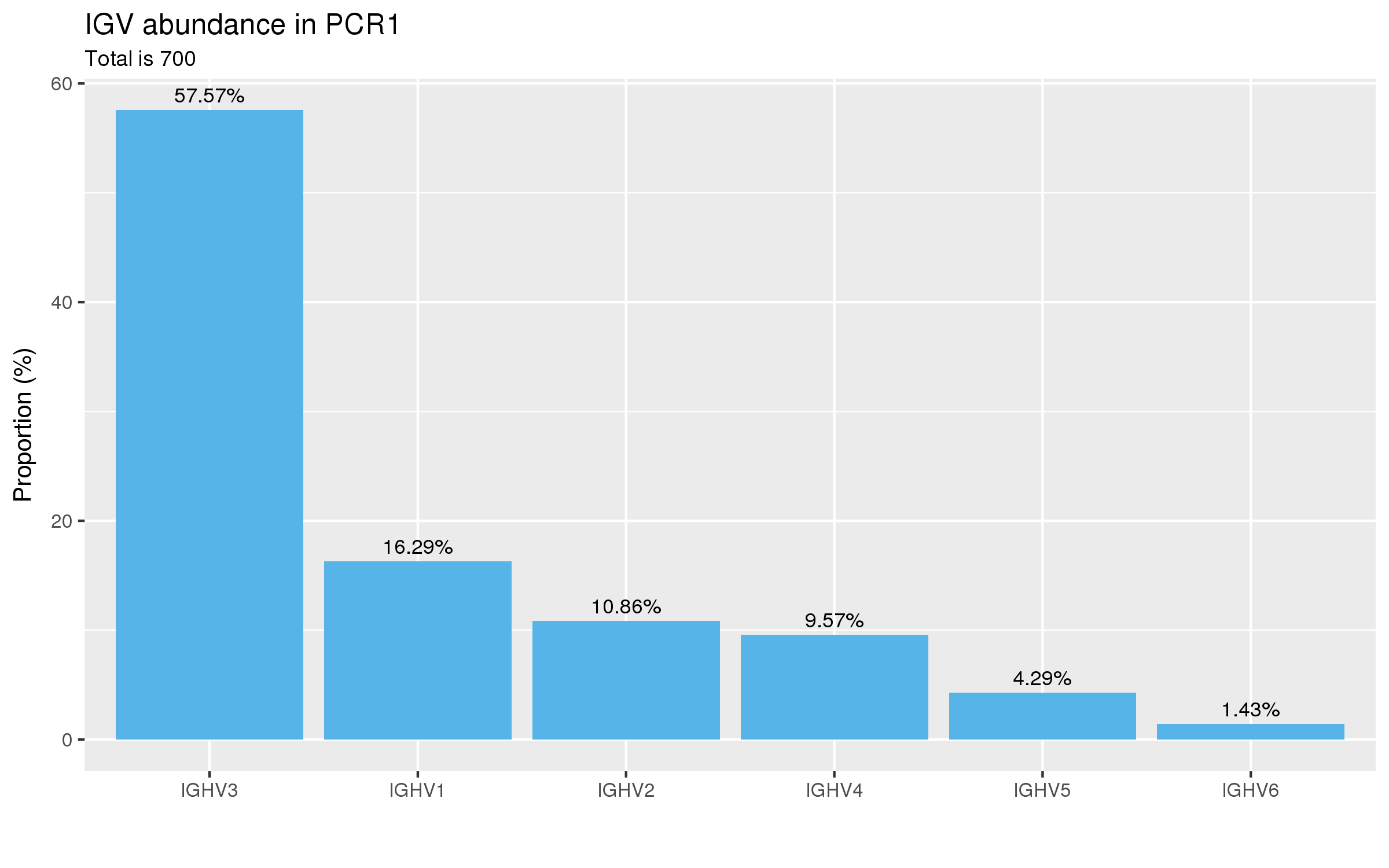 |
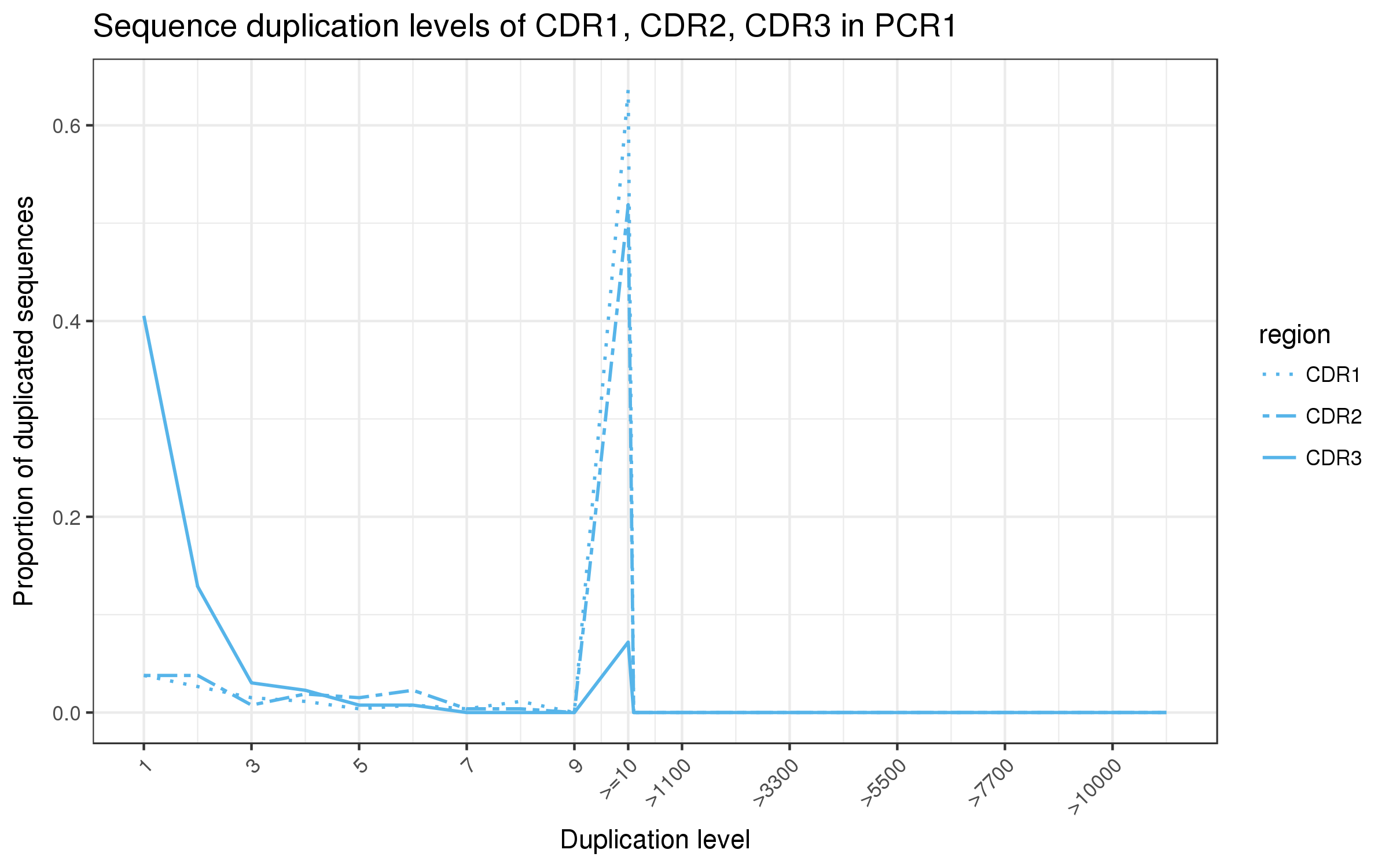 |
| Productivity analysis | Clonotype analysis |
|---|---|
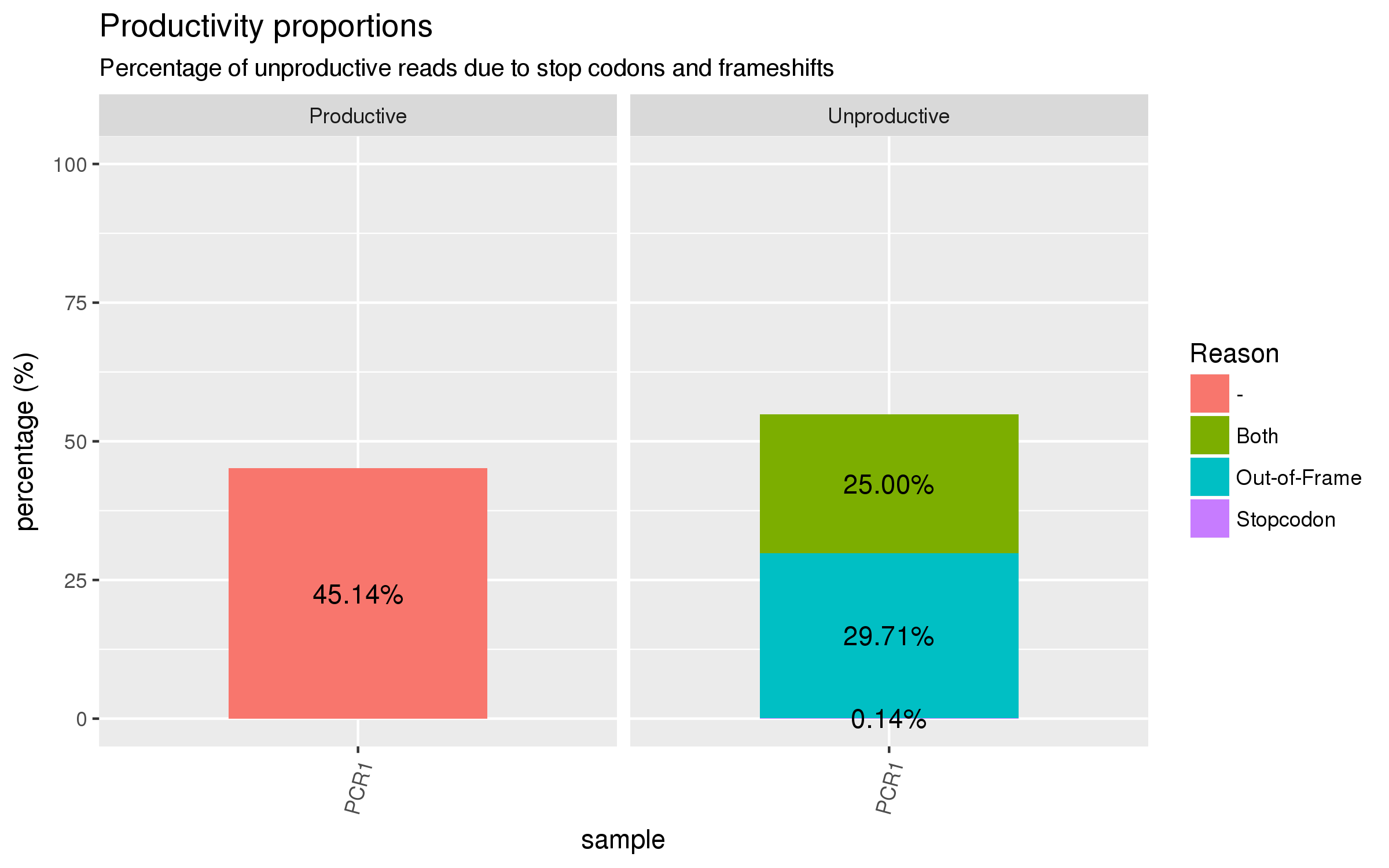 |
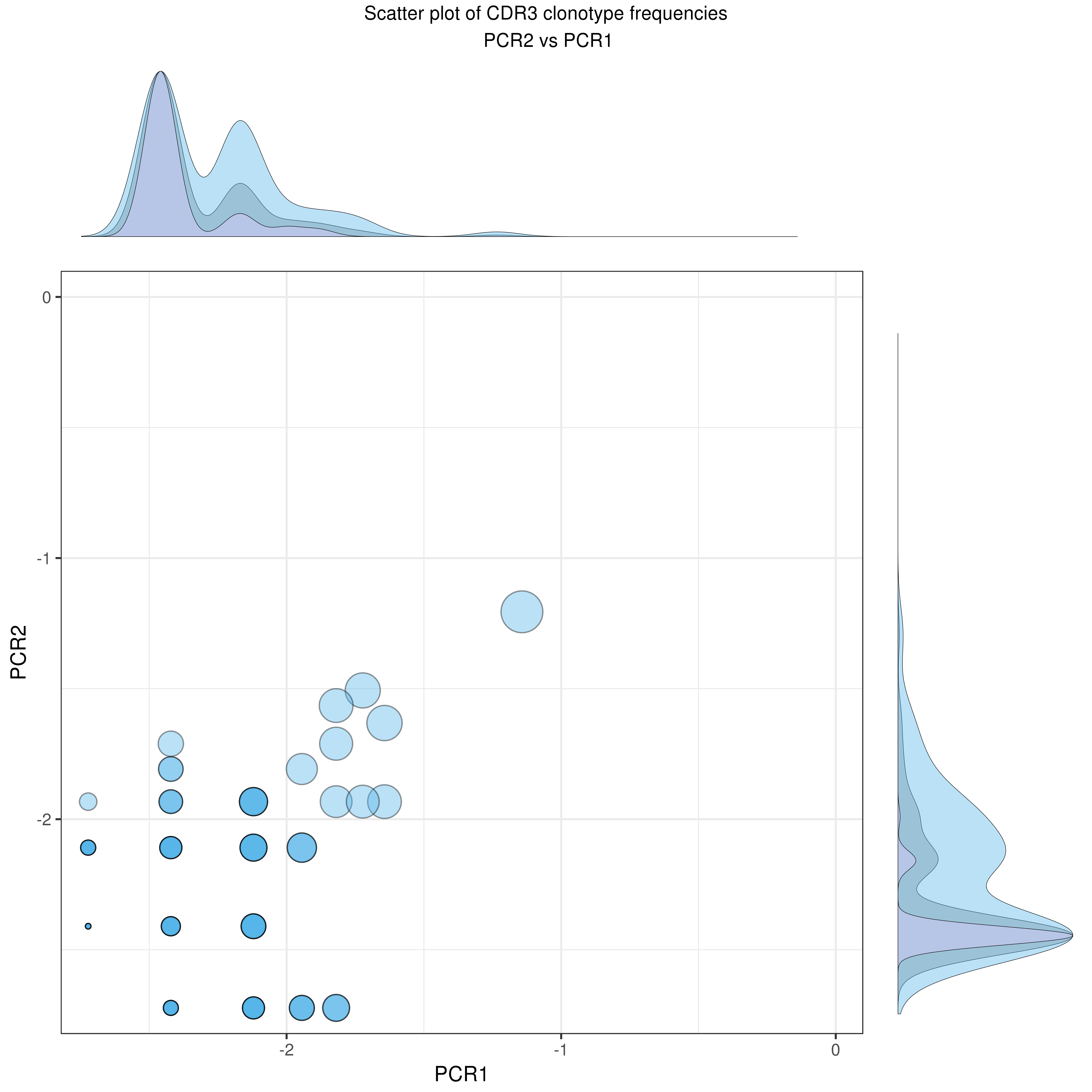 |
Additional plots along with documentation can be found in abseqR's main vignette.
In R console, type:
browseVignettes("abseqR")