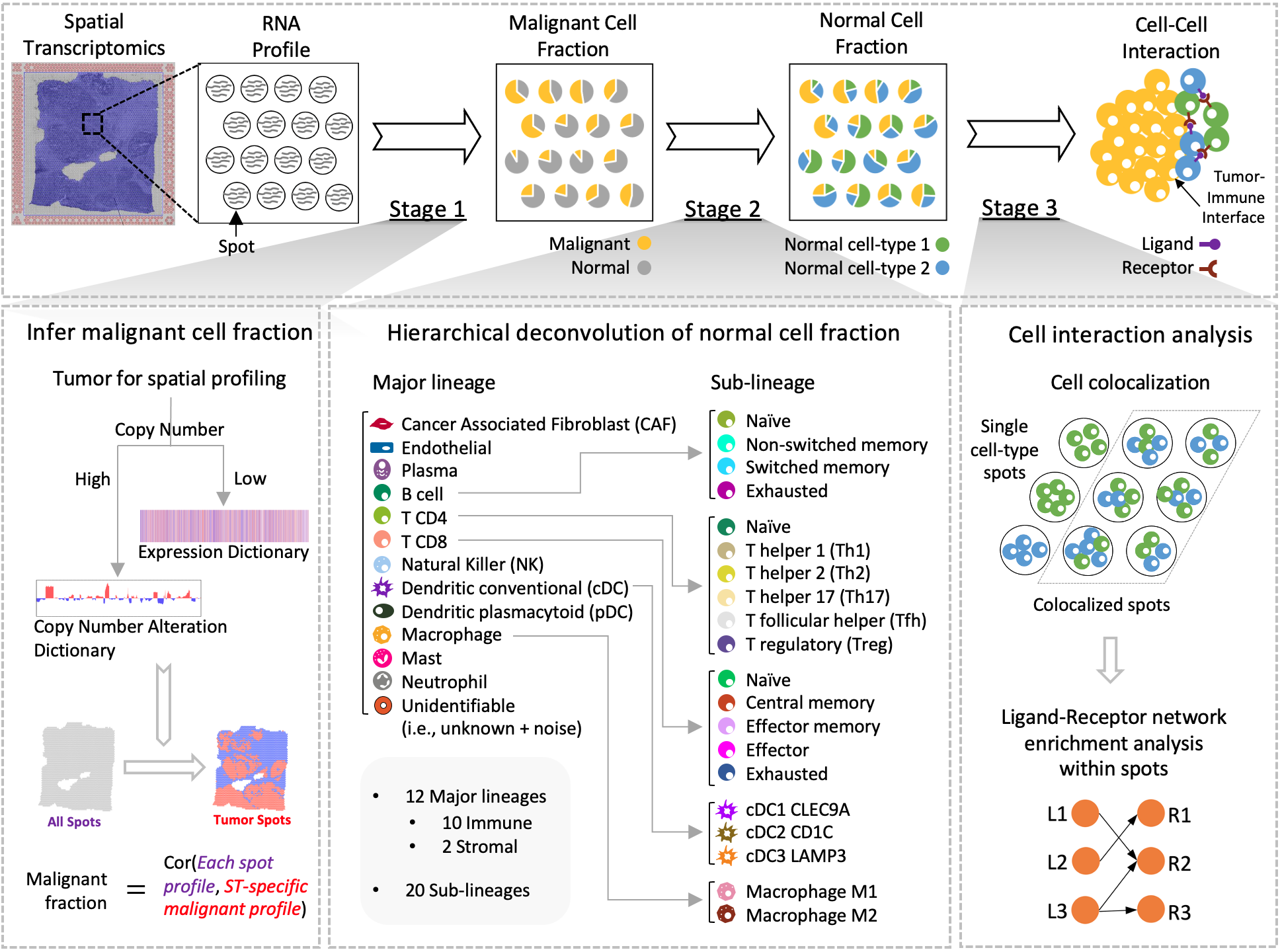
SpaCET is an R package desighed for analyzing cancer spatial transcriptomics (ST) datasets to estimate cell lineages and intercellular interactions within the tumor microenvironment. In a nutshell, SpaCET first infers cancer cell abundance by integrating a gene pattern dictionary of common malignancies. Subsequently, SpaCET employs a constrained linear regression model to calibrate local tissue densities and determine stromal and immune cell lineage fractions based on a comprehensive non-malignant cell atlas. Furthermore, SpaCET has the capability to unveil putative cell-cell interactions within the tumor microenvironment, particularly at the tumor-immune interface. Of note, although SpaCET does not require any input cell references for the analysis of tumor ST data, SpaCET can still incorporate a matched scRNA-seq dataset as customized references to conduct cell type deconvolution of any ST dataset.
To install SpaCET, we recommend using devtools:
# install.packages("devtools")
devtools::install_github("data2intelligence/SpaCET")Or user can install SpaCET from the source code. Click
here
to download it.
# install SpaCET in the R environment.
install.packages("Path_to_the_source_code", repos = NULL, type="source")- R version >= 4.2.0.
- R packages: Matrix, jsonlite, ggplot2, reshape2, patchwork, png, shiny, plotly, DT, MUDAN, factoextra, NbClust, cluster, parallel, psych, BiRewire, limma, UCell.
library(SpaCET)
visiumPath <- file.path(system.file(package = "SpaCET"), "extdata/Visium_BC")
SpaCET_obj <- create.SpaCET.object.10X(visiumPath = visiumPath)
SpaCET_obj <- SpaCET.deconvolution(SpaCET_obj, cancerType="BRCA", coreNo=8)
SpaCET_obj@results$deconvolution$propMat[1:13,1:5]
## 50x102 59x19 14x94 47x13 73x43
## Malignant 2.860636e-01 1 6.845966e-02 3.899756e-01 9.608802e-01
## CAF 3.118545e-01 0 3.397067e-01 1.111980e-01 3.372692e-02
## Endothelial 5.510895e-02 0 1.427060e-01 3.080531e-02 5.263544e-03
## Plasma 2.213392e-02 0 1.507382e-02 1.183170e-02 9.071809e-06
## B cell 3.885793e-03 0 9.271616e-02 1.406470e-01 1.329085e-06
## T CD4 1.344389e-01 0 1.554305e-02 1.249414e-01 1.112392e-05
## T CD8 7.578696e-03 0 2.514558e-07 1.379856e-03 1.123043e-06
## NK 7.104005e-04 0 1.670019e-06 4.890387e-08 3.562557e-07
## cDC 1.421632e-07 0 8.278023e-02 7.584295e-02 2.851146e-07
## pDC 1.606443e-06 0 2.283754e-02 1.805671e-02 3.878344e-07
## Macrophage 1.703304e-01 0 5.021248e-02 9.531511e-02 9.253645e-07
## Mast 7.905067e-08 0 1.621498e-05 1.333430e-07 1.162099e-07
## Neutrophil 1.380073e-05 0 9.528996e-07 1.167503e-08 9.908635e-05- Cell type deconvolution and interaction analysis
- Deconvolution with a matched scRNA-seq data set
- Gene set score calculation for spatial spots
Beibei Ru, Jinlin Huang, Yu Zhang, Kenneth Aldape, Peng Jiang. Estimation of cell lineages in tumors from spatial transcriptomics data. Nature Communications 14, 568 (2023). [Link]