It is an open-source global effort with collaborators from academia, biotech startups and big pharma.
Our goal is to improve ADMET (Absorption, Distribution, Metabolism, and Excretion) prediction tools.
We welcome everyone with the expertise in the field. If you're interested in collaboration, contact us via email or LinkedIn.
Our goal is to provide a tool that is:
- Accurate: It has higher characteristics compared to other open-source tools such as ADMETLab, Chemprop, QikProp etc.
- Open-source: The source code is freely available for anyone to view, use, modify and distribute.
- With simple API: It has an easy-to-use interface and can be integrated into various applications and platforms.
- Reproducible: You can access data sources, modeling workflow notebooks and models to easily reproduce and verify the entire modeling process.
- Easily deployable: It is easy to set up and use.
- Performant: It offers a reliable and high-performance solution for datasets of all sizes.
Our tool easily works with many platforms and applications. Here's what you can do with it when using Datagrok:
-
Mix your data: Combine your data with Datagrok's collected experimantal data for customized predictive models.
-
Visualize results: Use Datagrok tools for better predictions, data analysis and data visualization.
Currently, we have a total of 32 predictive models developed for Absorption, Distribution, Metabolism, Excretion and Toxicity.
| Name | Model | Size | Specificity | Sensitivity | Accuracy | Balanced Accuracy | ROC AUC |
|---|---|---|---|---|---|---|---|
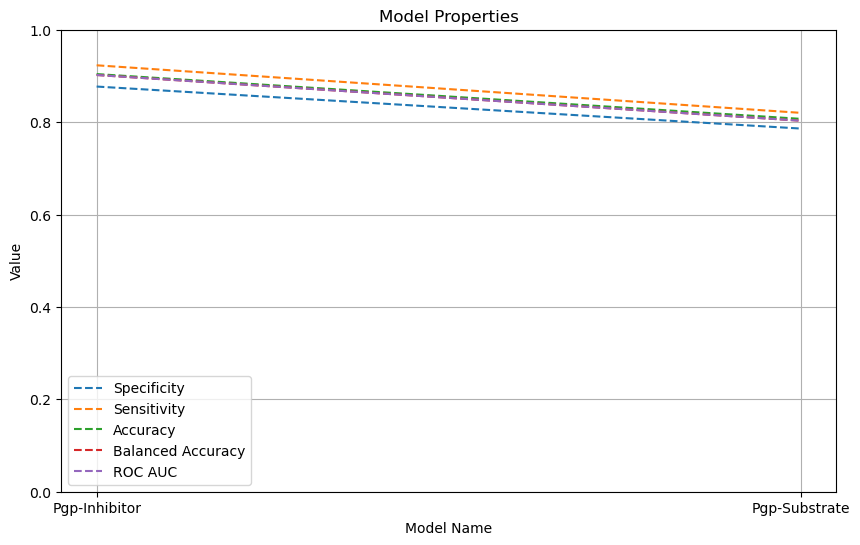
| Pgp-Inhibitor | Chemprop | 1,275 | 0.877 | 0.923 | 0.904 | 0.902 | 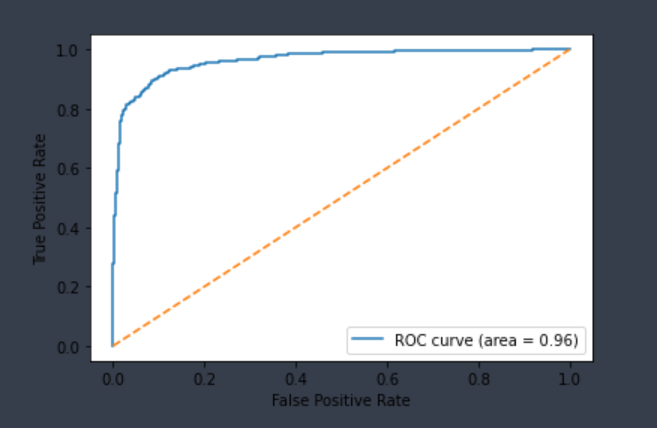 |
| Pgp-Substrate | Chemprop | 332 | 0.786 | 0.820 | 0.807 | 0.803 | 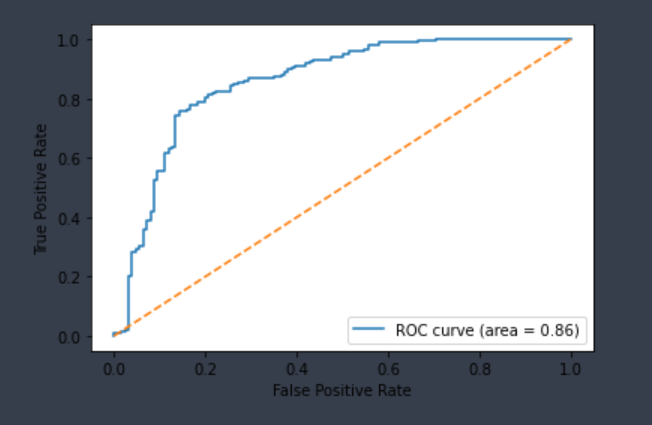 |
Below is a line chart that visually represents various metrics for the respective models.
| Name | Model | Size | MAE | RMSE | R2 | Spearman | True vs. Predicted | True vs. Residuals |
|---|---|---|---|---|---|---|---|---|
| PPBR | Chemprop | 2790 | 7.945 | 11.642 | 0.410 | 0.650 |  |
 |
| VDss | Chemprop | 1130 | 3.100 | 5.232 | 0.069 | 0.500 |  |
 |
| Name | Model | Size | Specificity | Sensitivity | Accuracy | Balanced Accuracy | ROC AUC |
|---|---|---|---|---|---|---|---|
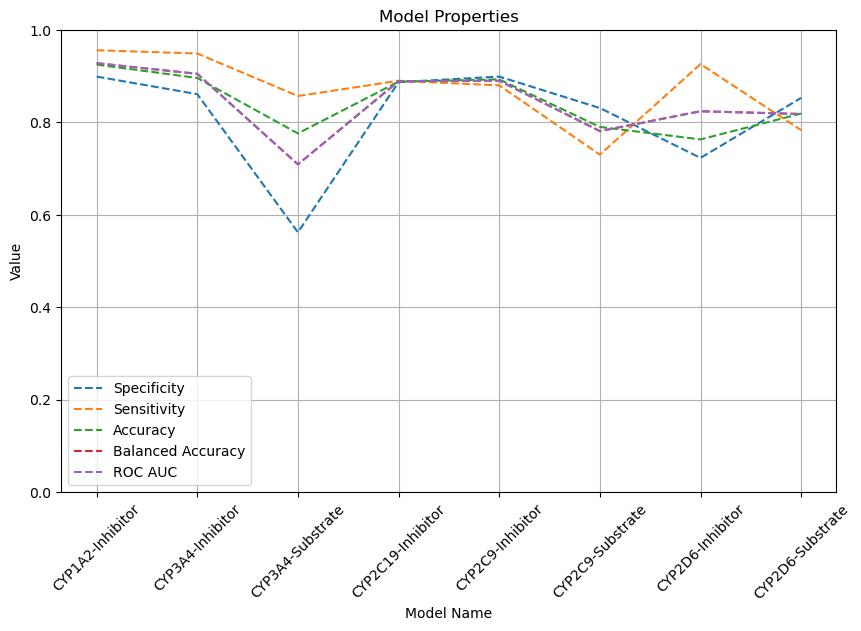
| CYP1A2-Inhibitor | Chemprop | 13,239 | 0.899 | 0.956 | 0.925 | 0.928 | 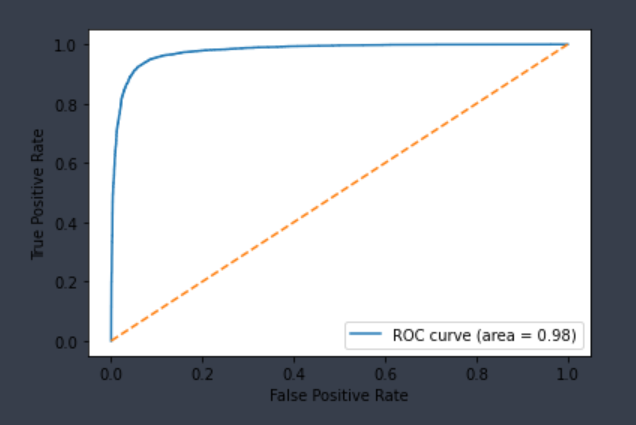 |
| CYP3A4-Inhibitor | Chemprop | 12,997 | 0.861 | 0.949 | 0.896 | 0.905 | 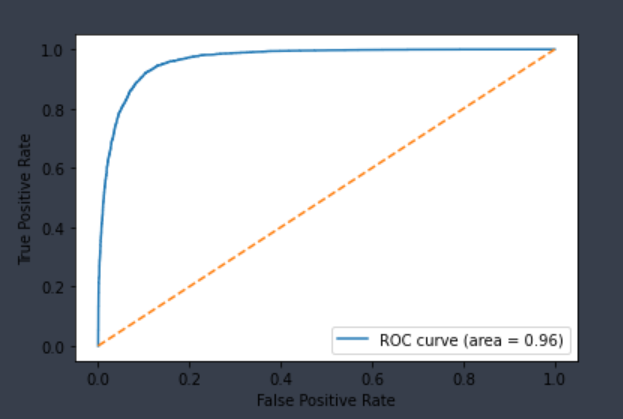 |
| CYP3A4-Substrate | Chemprop | 1,149 | 0.562 | 0.857 | 0.776 | 0.709 | 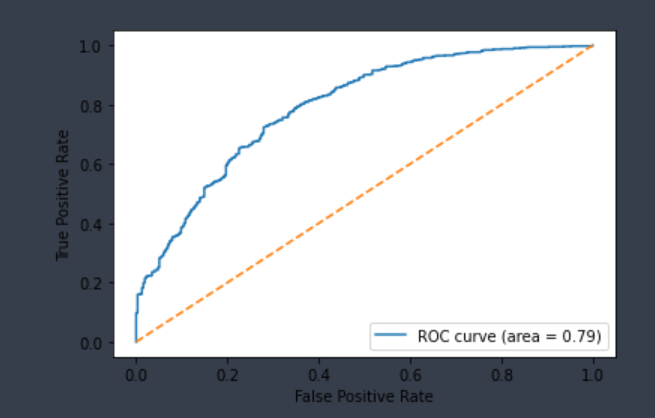 |
| CYP2C19-Inhibitor | Chemprop | 13,427 | 0.887 | 0.890 | 0.888 | 0.889 | 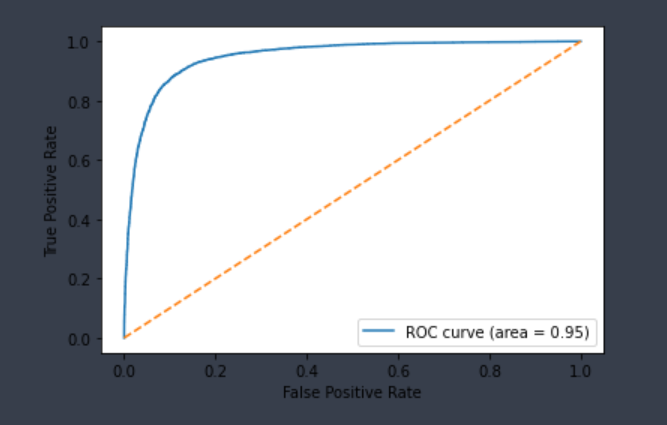 |
| CYP2C9-Inhibitor | Chemprop | 12,881 | 0.899 | 0.880 | 0.893 | 0.890 | 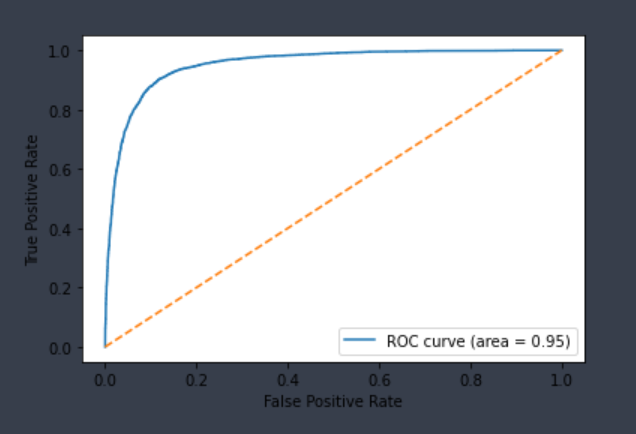 |
| CYP2C9-Substrate | Chemprop | 899 | 0.831 | 0.730 | 0.790 | 0.781 | 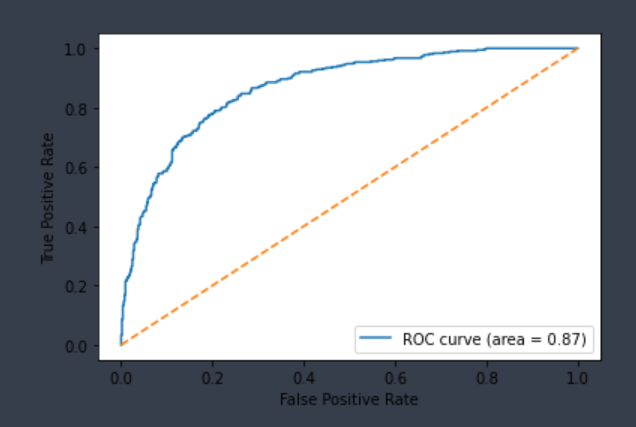 |
| CYP2D6-Inhibitor | Chemprop | 11,127 | 0.723 | 0.926 | 0.763 | 0.824 | 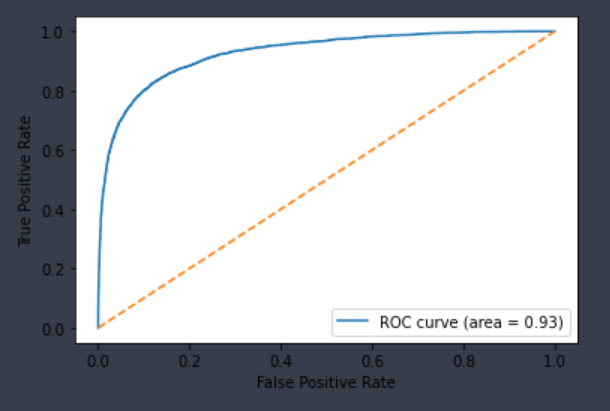 |
| CYP2D6-Substrate | Chemprop | 941 | 0.853 | 0.783 | 0.819 | 0.818 | 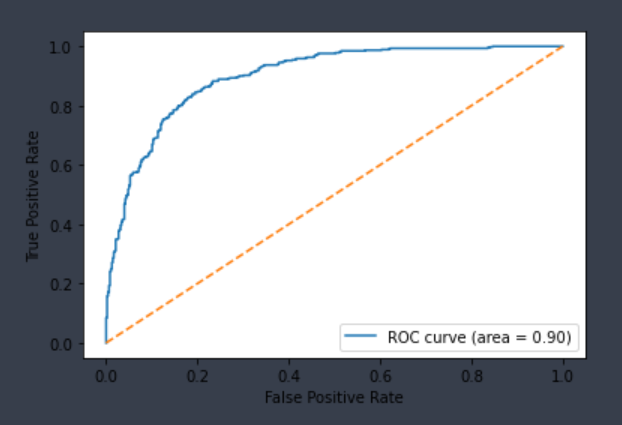 |
Here is a line chart illustrating various metrics for each of the corresponding models.
Admetica is a powerful tool for making ADMET predictions and can be easily installed on any operating system. You can install it using pip, and optionally, you can set up a conda environment for better package management.
To create a new conda environment and install Admetica, use the following commands:
conda create --name admetica-env python=3.11
conda activate admetica-envTo install Admetica, run:
pip install admetica==1.3By default, the pip installation will include all necessary dependencies for making ADMET predictions.
Admetica provides a command-line interface to make predictions. To use it, run:
admetica_predict \
--dataset-path data.csv \
--smiles-column smiles \
--properties Caco2,PPBR \
--save-path predictions.csvThis command assumes the presence of a file named data.csv with SMILES strings in the column smiles. In addition, you should specify the properties to be calculated (e.g. Caco2). The predictions will be saved to predictions.csv.
All models available in the repository are included and can be used.
In order to train a model or obtain predictions, you must provide data containing molecules (as SMILES strings) and known target values.
The data used in this research and its overview can be found in the Datasets folder.
You can create a model on your own using chemprop module or use publicly available models that are licalted in the Models folder.
To load a trained model and make predictions, run all the commands specified in the Chemical Property Prediction and Evaluation.ipynb file.
Our project is about improving and combining existing solutions, not reinventing the wheel. Here is the list of resources we've investigated:
- ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database / Jie Dong, Ning-Ning Wang, Zhi-Jiang Yao та ін. // J Cheminform. – 2018. – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6020094/.
- Evaluation of Free Online ADMET Tools for Academic or Small Biotech Environments / Júlia Dulsat, Blanca López-Nieto, Roger Estrada-Tejedor, José I. Borrell // Molecules. – 2023. – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9864198/.
- Vishwesh Venkatraman. FP-ADMET: a compendium of fingerprint-based ADMET prediction models / Vishwesh Venkatraman // J Cheminform. – 2021. – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8479898/.
- Front Pharmacol. vNN Web Server for ADMET Predictions / Front Pharmacol // Front Pharmacol. – 2017. – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5722789/.
- ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties / Guoli Xiong, Zhenxing Wu, Jiacai Yi та ін. // Nucleic Acids Res. – 2021. – https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8262709/.
- In silico Prediction of Chemical Ames Mutagenicity / Congying Xu, Feixiong Cheng, Lei Chen та ін. // J Cheminform. – 2012. – https://pubs.acs.org/doi/abs/10.1021/ci300400a.
- Computational Models for Human and Animal Hepatotoxicity with a Global Application Scope / Denis Mulliner, Friedemann Schmidt, Manuela Stolte та ін. // Chem. Res. Toxicol.. – 2016. – https://pubs.acs.org/doi/10.1021/acs.chemrestox.5b00465.
- ADMET Evaluation in Drug Discovery. 16. Predicting hERG Blockers by Combining Multiple Pharmacophores and Machine Learning Approaches / Shuangquan Wang, Huiyong Sun, Hui Liu та ін. // Mol. Pharmaceutics. – 2016. – https://pubs.acs.org/doi/10.1021/acs.molpharmaceut.6b00471.