Please navigate to this page for the original final report for the project:
https://datascisteven.github.io/Melanoma-Image-Classification
Presentation Link: https://prezi.com/view/JoLKnGuw0ZFBYFZra0c3/
While I had been exploring implementation through Flutter for app deployment, Flask seemed much more feasible given my time constraints and level of expertise.
The homepage asks for the user to upload a JPEG of any size into the application and to press SUBMIT once done.
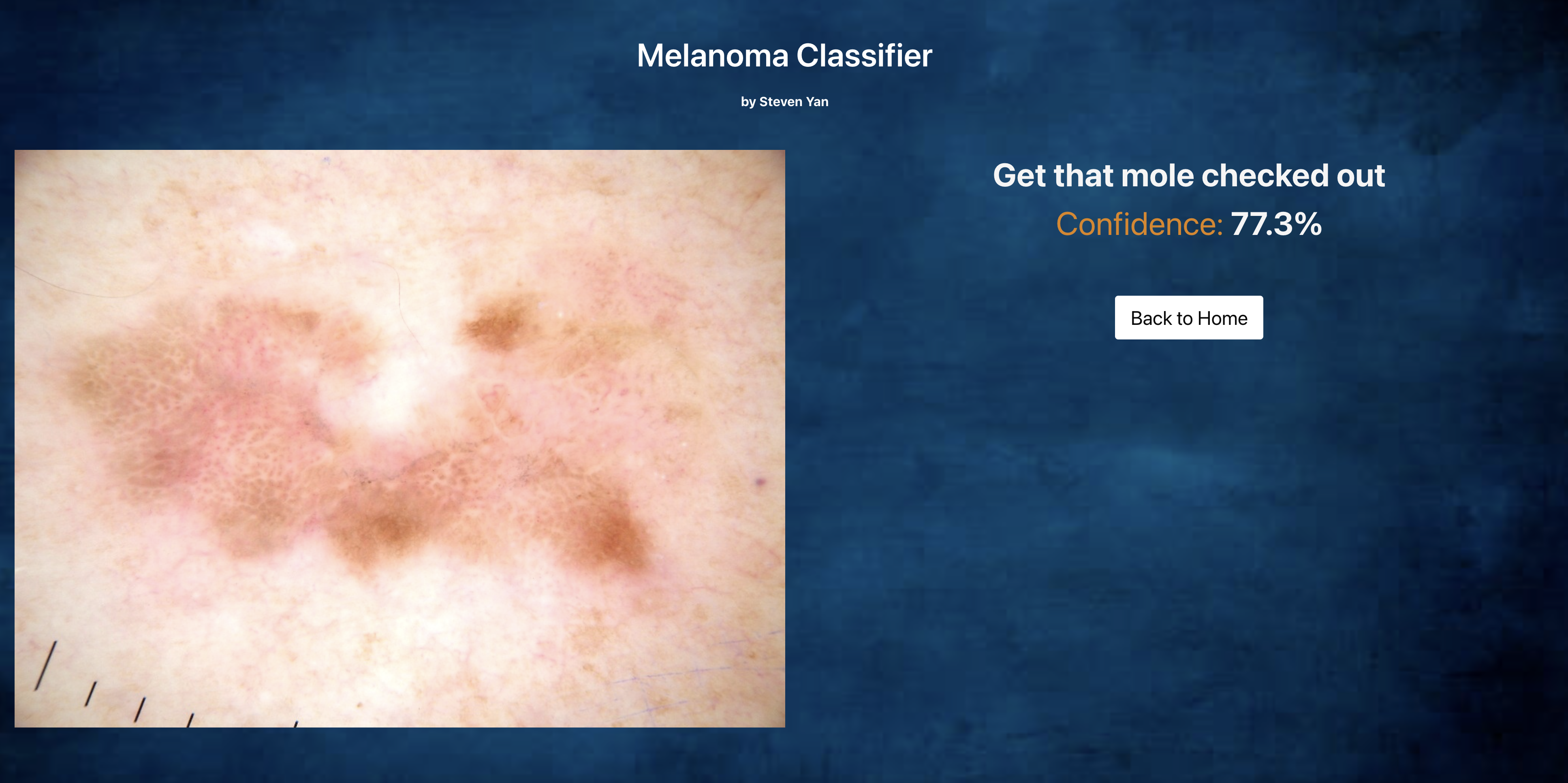
Upon pressing SUBMIT, you automatically get transferred to the Results page, and you are given a message to get the mole checked out or that it is just another beauty mark. The confidence level of that prediction is also given.
├── README.md <- the top-level README for reviewers of this project
├── _notebooks <- folder containing all the project notebooks
│ ├── albumentation.ipynb <- notebook for displaying augmentations
│ ├── EDA.ipynb <- notebook for dataset understanding and EDA
│ ├── folders.ipynb <- notebook for image folder management
│ ├── holdout.ipynb <- notebook for predicting on holdout sets
│ ├── preaugmentation.ipynb <- notebook for models with imbalanced dataset
│ ├── postaugmentation.ipynb <- notebook for models with dataset post-augmentations
│ ├── pretrained.ipynb <- notebook for pretrained models
│ └── utils.py <- py file with self-defined functions
├── final_notebook.ipynb <- final notebook for capstone project
├── _data <- folder of csv files (csv)
├── MVP Presentation.pdf <- pdf of the MVP presentation
├── _Melanoma-Flask <- folder with Flask application
└── utils.py <- py file with self-defined functions
Steven Yan
![]() Email: stevenyan@uchicago.edu
Email: stevenyan@uchicago.edu
![]() LinkedIn: https://www.linkedin.com/in/datascisteven
LinkedIn: https://www.linkedin.com/in/datascisteven
![]() Github: https://www.github.com/datascisteven
Github: https://www.github.com/datascisteven
International Skin Imaging Collaboration. SIIM-ISIC 2020 Challenge Dataset. International Skin Imaging Collaboration https://doi.org/10.34970/2020-ds01 (2020).
Rotemberg, V. et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 8: 34 (2021). https://doi.org/10.1038/s41597-021-00815-z
ISIC 2019 data is provided courtesy of the following sources:
- BCN20000 Dataset: (c) Department of Dermatology, Hospital Clínic de Barcelona
- HAM10000 Dataset: (c) by ViDIR Group, Department of Dermatology, Medical University of Vienna; https://doi.org/10.1038/sdata.2018.161
- MSK Dataset: (c) Anonymous; https://arxiv.org/abs/1710.05006 ; https://arxiv.org/abs/1902.03368
Tschandl, P. et al. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 5: 180161 doi: 10.1038/sdata.2018.161 (2018)
Codella, N. et al. “Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC)”, 2017; arXiv:1710.05006.
Marc Combalia, Noel C. F. Codella, Veronica Rotemberg, Brian Helba, Veronica Vilaplana, Ofer Reiter, Allan C. Halpern, Susana Puig, Josep Malvehy: “BCN20000: Dermoscopic Lesions in the Wild”, 2019; arXiv:1908.02288.
Codella, N. et al. “Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC)”, 2018; https://arxiv.org/abs/1902.03368