Official implementation of RadAdapt from Stanford University
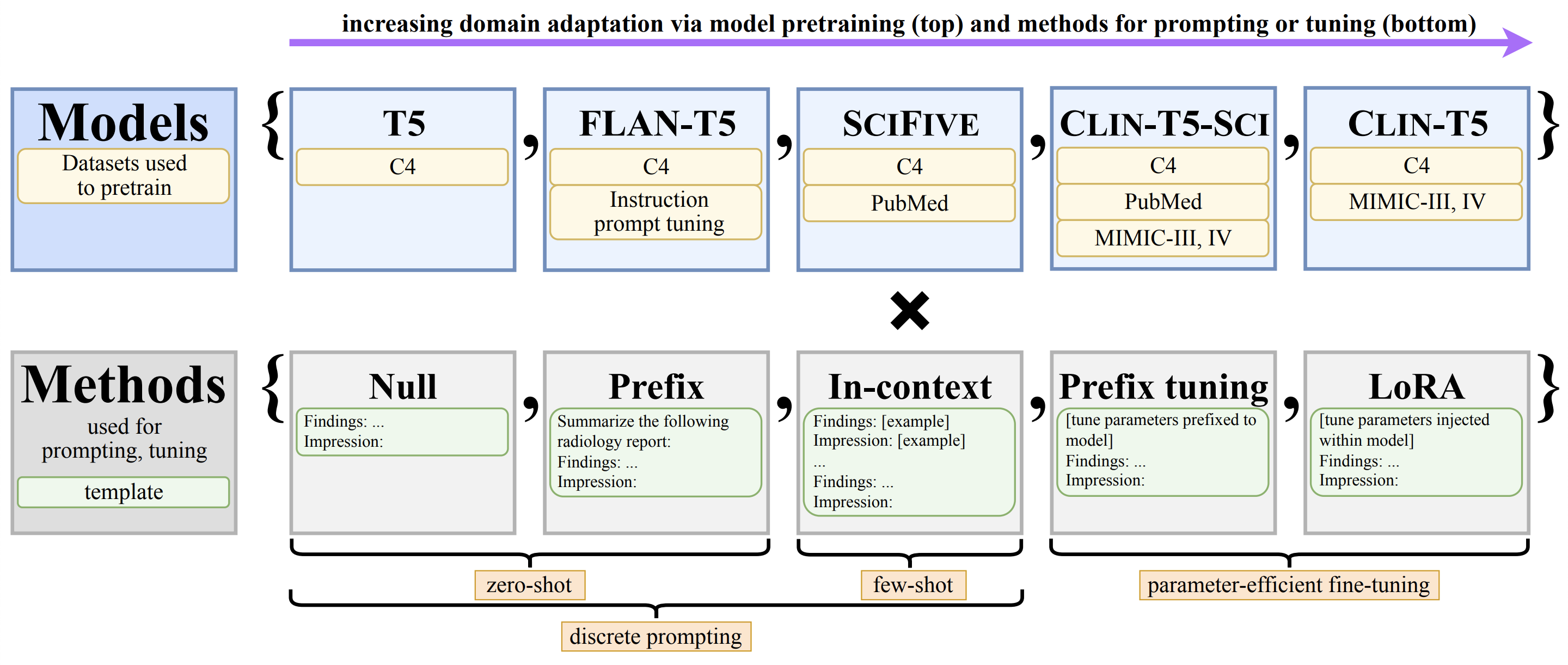
- Title: RadAdapt: Radiology Report Summarization via Lightweight
Domain Adaptation of Large Language Models
- Authors: Dave Van Veen*, Cara Van Uden*, Maayane Attias, Anuj Pareek, Christian Bluethgen, Malgorzata Polacin, Wah Chiu, Jean-Benoit Delbrouck, Juan Manuel Zambrano Chaves, Curtis P. Langlotz, Akshay S. Chaudhari, John Pauly
- Contact: {vanveen, cvanuden} [at] stanford [dot] edu
Use these commands to set up a conda environment:
conda env create -f env/environment.yml
conda activate radadapt
If your CUDA toolkit is older than 11.6 (display via nvcc --version), refer to env/README.md for modified instructions.
- In
src/constants.py, set your own project directoryDIR_PROJECT. - Run a script, setting
modelandcase_idas desired:run_discrete.sh: generate output via discrete prompting.train_peft.sh: fine-tune a model using a parameter-efficient method. LoRA is recommended.run_peft.sh: generate output from a fine-tuned model.calc_metrics.sh: calculate metrics on outputs.
- To modify default parameters, create a new
casesentry insrc/constants.py. - To add your own dataset, follow the format in
data/, which contains a subset of chest x-ray reports from Open-i.
@article{van2023radadapt,
title={RadAdapt: Radiology Report Summarization via Lightweight Domain Adaptation of Large Language Models},
author={Van Veen, Dave and Van Uden, Cara and Attias, Maayane and Pareek, Anuj and Bluethgen, Christian and Polacin, Malgorzata and Chiu, Wah and Delbrouck, Jean-Benoit and Chaves, Juan Manuel Zambrano and Langlotz, Curtis P and others},
journal={arXiv preprint arXiv:2305.01146},
year={2023}
}