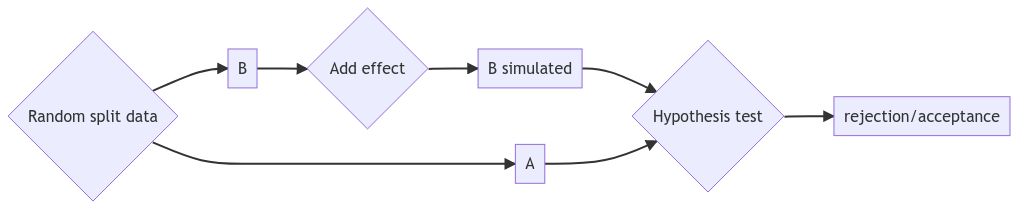
A library to run simulation-based power analysis, including cluster-randomized trial data. Also useful to design and analyse cluster-randomized and switchback experiments.
Hello world of the library, non-clustered version. There is an outcome variable analyzed with a linear regression. The perturbator adds a constant effect to treated units, and the splitter is random.
import numpy as np
import pandas as pd
from cluster_experiments import PowerAnalysis
# Create fake data
N = 1_000
df = pd.DataFrame(
{
"target": np.random.normal(0, 1, size=N),
}
)
config = {
"analysis": "ols_non_clustered",
"perturbator": "constant",
"splitter": "non_clustered",
"n_simulations": 50,
}
pw = PowerAnalysis.from_dict(config)
# Keep in mind that the average effect is the absolute effect added, this is not relative!
power = pw.power_analysis(df, average_effect=0.1)
# You may also get the power curve by running the power analysis with different average effects
power_line = pw.power_line(df, average_effects=[0, 0.1, 0.2])
# A faster method can be used to run the power analysis, using the approximation of
# the central limit theorem, which is stable with less simulations
from cluster_experiments import NormalPowerAnalysis
npw = NormalPowerAnalysis.from_dict(
{
"analysis": "ols_non_clustered",
"splitter": "non_clustered",
"n_simulations": 5,
}
)
power_line_normal = npw.power_line(df, average_effects=[0, 0.1, 0.2])
# you can also use the normal power to get mde from a power level
mde = npw.mde(df, power=0.8)Hello world of this library, clustered version. Since it uses dates as clusters, we consider it a switchback experiment. However, if you want to run a clustered experiment, you can use the same code without the dates.
from datetime import date
import numpy as np
import pandas as pd
from cluster_experiments.power_analysis import PowerAnalysis
# Create fake data
N = 1_000
clusters = [f"Cluster {i}" for i in range(100)]
dates = [f"{date(2022, 1, i):%Y-%m-%d}" for i in range(1, 32)]
df = pd.DataFrame(
{
"cluster": np.random.choice(clusters, size=N),
"target": np.random.normal(0, 1, size=N),
"date": np.random.choice(dates, size=N),
}
)
config = {
"cluster_cols": ["cluster", "date"],
"analysis": "gee",
"perturbator": "constant",
"splitter": "clustered",
"n_simulations": 50,
}
pw = PowerAnalysis.from_dict(config)
print(df)
# Keep in mind that the average effect is the absolute effect added, this is not relative!
power = pw.power_analysis(df, average_effect=0.1)
print(f"{power = }")This is a more comprehensive example of how to use this library. There are simpler ways to run this power analysis above but this shows all the building blocks of the library.
from datetime import date
import numpy as np
import pandas as pd
from cluster_experiments.experiment_analysis import GeeExperimentAnalysis
from cluster_experiments.perturbator import ConstantPerturbator
from cluster_experiments.power_analysis import PowerAnalysis, NormalPowerAnalysis
from cluster_experiments.random_splitter import ClusteredSplitter
# Create fake data
N = 1_000
clusters = [f"Cluster {i}" for i in range(100)]
dates = [f"{date(2022, 1, i):%Y-%m-%d}" for i in range(1, 32)]
df = pd.DataFrame(
{
"cluster": np.random.choice(clusters, size=N),
"target": np.random.normal(0, 1, size=N),
"date": np.random.choice(dates, size=N),
}
)
# A switchback experiment is going to be run, prepare the switchback splitter for the analysis
sw = ClusteredSplitter(
cluster_cols=["cluster", "date"],
)
# We use a constant perturbator to add artificial effect on the treated on the power analysis
perturbator = ConstantPerturbator()
# Use gee to run the analysis
analysis = GeeExperimentAnalysis(
cluster_cols=["cluster", "date"],
)
# Run the power analysis
pw = PowerAnalysis(
perturbator=perturbator, splitter=sw, analysis=analysis, n_simulations=50, seed=123
)
# Keep in mind that the average effect is the absolute effect added, this is not relative!
power = pw.power_analysis(df, average_effect=0.1)
print(f"{power = }")
# You can also use normal power analysis, that uses central limit theorem to estimate power, and it should be stable in less simulations
npw = NormalPowerAnalysis(
splitter=sw, analysis=analysis, n_simulations=50, seed=123
)
power = npw.power_analysis(df, average_effect=0.1)
print(f"{power = }")The library offers the following classes:
- Regarding power analysis:
PowerAnalysis: to run power analysis on any experiment design, using simulationPowerAnalysisWithPreExperimentData: to run power analysis on a clustered/switchback design, but adding pre-experiment df during split and perturbation (especially useful for Synthetic Control)NormalPowerAnalysis: to run power analysis on any experiment design using the central limit theorem for the distribution of the estimator. It can be used to compute the minimum detectable effect (MDE) for a given power level.ConstantPerturbator: to artificially perturb treated group with constant perturbationsBinaryPerturbator: to artificially perturb treated group for binary outcomesRelativePositivePerturbator: to artificially perturb treated group with relative positive perturbationsNormalPerturbator: to artificially perturb treated group with normal distribution perturbationsBetaRelativePositivePerturbator: to artificially perturb treated group with relative positive beta distribution perturbationsBetaRelativePerturbator: to artificially perturb treated group with relative beta distribution perturbations in a specified support intervalSegmentedBetaRelativePerturbator: to artificially perturb treated group with relative beta distribution perturbations in a specified support interval, but using clusters
- Regarding splitting data:
ClusteredSplitter: to split data based on clustersFixedSizeClusteredSplitter: to split data based on clusters with a fixed size (example: only 1 treatment cluster and the rest in control)BalancedClusteredSplitter: to split data based on clusters in a balanced wayNonClusteredSplitter: Regular data splitting, no clustersStratifiedClusteredSplitter: to split based on clusters and strata, balancing the number of clusters in each stratusRepeatedSampler: for backtests where we have access to counterfactuals, does not split the data, just duplicates the data for all groups- Switchback splitters (the same can be done with clustered splitters, but there is a convenient way to define switchback splitters using switch frequency):
SwitchbackSplitter: to split data based on clusters and dates, for switchback experimentsBalancedSwitchbackSplitter: to split data based on clusters and dates, for switchback experiments, balancing treatment and control among all clustersStratifiedSwitchbackSplitter: to split data based on clusters and dates, for switchback experiments, balancing the number of clusters in each stratus- Washover for switchback experiments:
EmptyWashover: no washover done at all.ConstantWashover: accepts a timedelta parameter and removes the data when we switch from A to B for the timedelta interval.
- Regarding analysis:
GeeExperimentAnalysis: to run GEE analysis on the results of a clustered designMLMExperimentAnalysis: to run Mixed Linear Model analysis on the results of a clustered designTTestClusteredAnalysis: to run a t-test on aggregated data for clustersPairedTTestClusteredAnalysis: to run a paired t-test on aggregated data for clustersClusteredOLSAnalysis: to run OLS analysis on the results of a clustered designOLSAnalysis: to run OLS analysis for non-clustered dataTargetAggregation: to add pre-experimental data of the outcome to reduce varianceSyntheticControlAnalysis: to run synthetic control analysis
- Other:
PowerConfig: to conviently configurePowerAnalysisclass
You can install this package via pip.
pip install cluster-experimentsIt may be safer to install via;
python -m pip install cluster-experimentsIn case you want to use venv as a virtual environment:
python -m venv venv
source venv/bin/activateAfter creating the virtual environment (or not), run:
git clone git@github.com:david26694/cluster-experiments.git
cd cluster-experiments
make install-dev