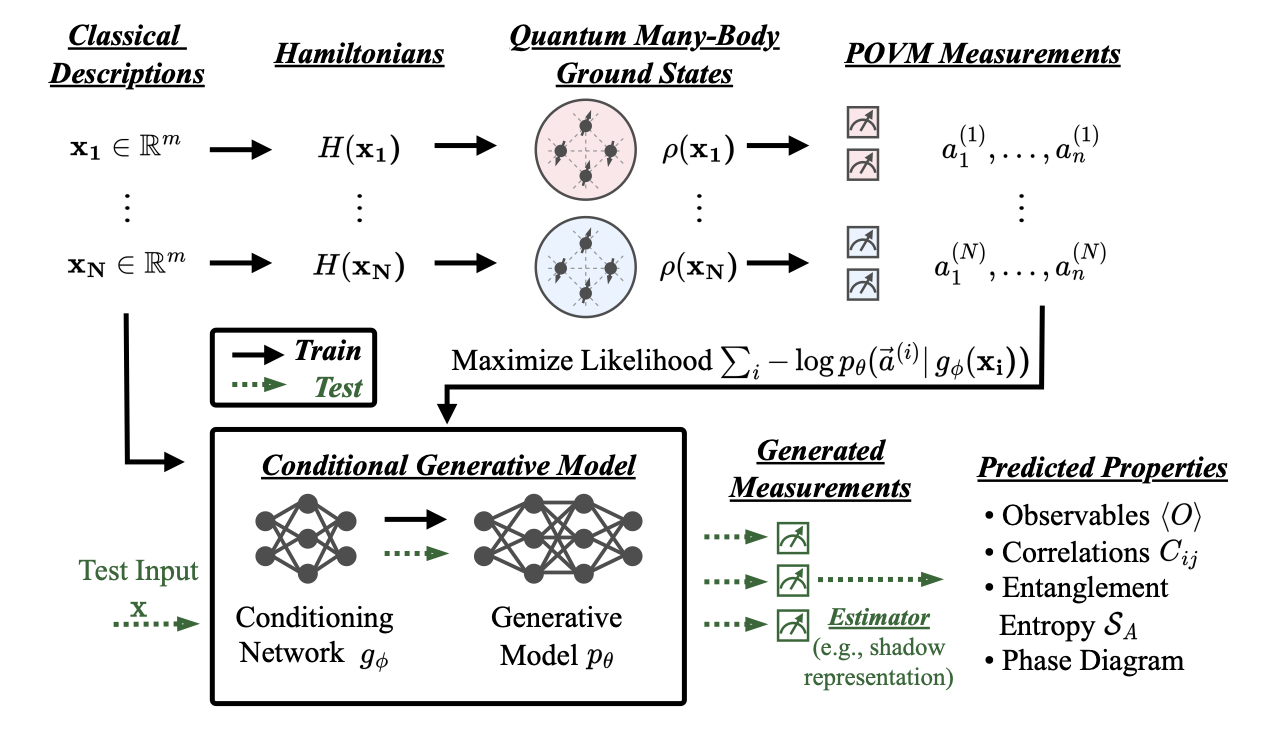
This repo contains the code accompanying the paper "Predicting Properties of Quantum Systems with Conditional Generative Models" [arXiv].
If you find this repo useful for your research, please consider citing our paper:
@article{cond-generative-quantum-states,
title={Predicting Properties of Quantum Systems with Conditional Generative Models},
author={Wang, Haoxiang and Weber, Maurice and Izaac, Josh and Lin, Cedric Yen-Yu},
journal={arXiv preprint arXiv:2211.16943},
year={2022},
}- Python (tested with python 3.8 & 3.9)
- Please see
requirements.txtfor the required Python packages (if you are using pip or Conda, you can runpip install -r requirements.txt) - Pytorch-Geometric (PyG): Please refer to
its official documentation for
installation.
- PyG is used in our experiments on 2D Heisenberg models (our conditional generative model has a GNN module in that case).
- Jax: Please refer to its official documentation for installation (a
CPU version is sufficient).
jaxandneural-tangents(a Jax-based package) are only used when comparing our method with Neural Tangent Kernel (used by Hsin-Yuan Huang et al. in their Science 2022 paper). You can skip this installation if you do not intend to do the comparison.
- Please see
- Julia (tested with version 1.7 & 1.8)
- The Julia language can be downloaded from the official website (If you are using pip or Conda as your python package manager, you can also install Julia with the Jill package).
- Run
julia install_pacakges.jlin therydberg/folder to install all necessary Julia packages.- If you want to use GPU for simulation, you need to install the
CUDAandAdaptpackages in addition ( seerydberg/README.mdfor details).
- If you want to use GPU for simulation, you need to install the
-
To train the models, we recommend using a GPU with at least 16GB memory (otherwise, you may need to use a smaller batch size).
-
For inference with our pre-trained models, a CPU is sufficient (may need to decrease the batch size to accommodate the memory).
Download - All simulation data & pre-trained models can be downloaded from this Google Drive
folder.
It contains two sub-folders, data/ and logs/. Please put them in the root of this repo after downloading.
In the paper, we use the following datasets:
- Heisenberg Models (
data/2d_heisenberg_data/)- Our simulated data with PennyLane (generated using our
code
heisenberg_generate_data.py) - Hsin-Yuan Huang's simulated data, available in this GitHub repo.
- Our simulated data with PennyLane (generated using our
code
- Rydberg Atom Systems (
data/rydberg/)- Our simulated data via Bloqade.jl (generated using our code
in
rydberg)- 1D Rydberg-atom chains of 13,15,...,33 atoms.
- 2D Rydberg-atom square lattices of 5x5 atoms (with various adiabatic evolution times for the ground-state preparation).
- Our simulated data via Bloqade.jl (generated using our code
in
First clone the provable-ml-quantum repo. Since our scripts
expect the data to be organized slightly differently, and to create train and test splits, we provide the
reorganize_heisenberg_data.py script. Run it as follows:
python reorganize_heisenberg_data.py <path/to/provable-ml-quantum/heisenberg/data>This will reorganize the data and save it to data/2d_heisenberg_data/{rows}x{cols}/{split}/.
We trained conditional transformers over the simulation data. If you want to directly load pre-trained models when using
our scripts, please put them in logs/.
In Sec. III-A of the paper, we conducted various numerical experiments over Heisenberg models of various sizes.
The trained models are saved in logs/2d_heisenberg_checkpoints/.
In Sec. III-B of the paper, we conducted 4 machine learning experiments over Rydberg atom systems, and we provide
pre-trained models for them in different sub-folders. Notice that we trained models for a different number of
iterations, and marked them with suffixes like ...{#iterations}.pth (e.g., ...100k.pth implies training with 100k
iterations).
- Sec. III-B(1) - Predicting quantum phases
logs/rydberg_1D/: models trained on 1D Rydberg-atom chains of 31 atoms.logs/rydberg_2D/: models trained on 2D Rydberg-atom chains of 25 atoms (prepared with adiabatic evolution of 3.0 μs)
- Sec. III-B(2) - Predicting phases of larger quantum systems
logs/rydberg_1D-size/: models trained on 1D Rydberg-atom chains of 13,15,...,27 atoms.
- Sec. III-B(3) - Predicting phases of ground states prepared with longer adiabatic evolution time
logs/rydberg_2D-time/: models trained on 2D Rydberg-atom square lattices of 5x5 atoms, which are prepared with adiabatic evolution of 0.4, 0.6, 0.8, 1.0 μs.
In this repo, we also provide tutorials that are intended to communicate the ideas of our paper in a more interactive manner.
-
Heisenberg Models. The notebook
Tutorial-2D-Heisenberg.ipynbfirst introduces the 2D random Heisenberg model, and shows how correlations and entanglement entropies of its ground states can be calculated. In the second part, we show how a pretrained conditional generative model can be used to generate samples corresponding to a ground state of a random Heisenberg Hamiltonian. Finally, in the last part, we walk you through the code used to train a transformer model, conditioned on the coupling graph, which was embedded by a graph convolutional neural network. -
Rydberg Atom Systems. The notebook
Tutorial-Rydberg-1D.ipynbintroduces 1D Rydberg atom systems, loads simulation data of such systems with 31 atoms (generated via Julia inrydberg/), and plots the phase diagram of 1D Rydberg-atom chains based on the simulation data. Then, we demonstrate how to train a conditional generative model over a subset of the simulation data (you can also directly load a pre-trained model fromlogs/rydberg_1D/). With the trained model, you can generate measurements for any new Rydberg-atom system of the same kind, and predict its phases (Disordered,$Z_2$ -ordered, or$Z_3$ -ordered). In this tutorial, we use the trained model to predict the entire phase diagram of 1D Rydberg-atom chains (31 atoms), and compare it with the predicted phase diagrams by several kernel methods (introduced by Hsin-Yuan Huang et al. in their Science 2022 paper) - you can see that the prediction by our method is much more accurate that these kernel methods.
- Generate your own data
You can generate your own dataset consisting of randomized Pauli measurements using the
script generate_heisenberg_data.py.
The script first samples random coupling matrices of the specified dimensions, calculates the corresponding ground state
using exact diagonalization and then samples random Pauli measurements using the
Classical Shadow implementation in PennyLane.
For example, if you want to create a dataset for a 4x4 lattice, with 40 train Hamiltonians and 10 test Hamiltonians,
with 500 measurements for each Hamiltonian, you can run
python heisenberg_generate_data.py --nh_train 40 --nh_test 10 --rows 4 --cols 4 --shots 500 The resulting data will be saved under data/2d_heisenberg/4x4 with two sub-folders train and test for the
respective data splits.
- Open source data
For systems with 20 and more qubits, we have used publicly available data from this GitHub repo by Hsin-Yuan Huang. These datasets were simulated using DMRG. Or, you can directly download the data from our Google Drive folder.
To train a conditional generative model for the 2D anti-ferromagnetic random Heisenberg model, you can use the
script heisenberg_train_transformer.py. An example command is
python heisenberg_train_transformer.py --train-size 4x4 --train-samples 500which uses the default hyperparameters used throughout the paper and trains a model on all the Hamiltonians
sampled in the previous step. Note that this is compute-intensive and should be run with GPU support.
If you want to quickly test on CPUs, you can set the flag --hamiltonians 1 to use data from only a single Hamiltonian.
To generate samples from a trained conditional generative model, you can use the script
heisenberg_sample_transformer.py. The flag --results-dir indicates the directory pointing to the run package where
the results of a training run have been saved. Note that this should be the root of the results directory and not the
checkpoints folder, as this script requires access to flags set during training and saved to the args.json file.
Evaluating properties of ground states of the Heisenberg model can be done using the script
heisenberg_evaluate_properties.py. Note that this requires you to have completed the previous steps and generated
samples from a trained model using the script heisenberg_sample_transformer.py. Similar to the previous step, the flag
--results-root indicates the directory pointing to the run package where the results of a training run have been
saved.
The flag --snapshots indicates the number of samples (i.e., snapshots) which will be used to estimate the correlation
functions and the entanglement entropies. We use the classical shadow implementation in PennyLane to compute these
properties.
You can generate your own dataset using our Julia simulation code in rydberg/. Or, you can download the data from
this Google Drive folder.
To reproduce our experiments on Rydberg atom systems, you can check out the following Jupyter notebooks (with detailed descriptions inside):
- Sec. III-B(1) - Predicting quantum phases
- 1D Lattices:
Tutorial-Rydberg-1D.ipynb - 2D Lattices:
notebooks/Experiment-Rydberg-2D.ipynb- Furthermore, we provide a demo,
notebooks/Demo-Rydberg-2D-Phases.ipynb, to visualize phases of 2D Rydberg-atom lattices (in both the real space and the Fourier space).
- Furthermore, we provide a demo,
- 1D Lattices:
- Sec. III-B(2) - Predicting phases of larger quantum systems
notebooks/Experiment-Rydberg-1D-LargerSystems.ipynb
- Sec. III-B(3) - Predicting phases of ground states prepared with longer adiabatic evolution time
notebooks/Experiment-Rydberg-2D-LongerEvolution.ipynb
We found several very helpful codebases when building this repo, and we sincerely thank their authors:
- Bloqade.jl: A Julia package for neutral-atom simulations.
- provable-ml-quantum: Official implementation for Provably efficient machine learning for quantum many-body problems.
- PennyLane Tutorials:
- Classical Shadows: A Python re-implementation of Hsin-Yuan Huang's classical shadows code (C++).
- Machine learning for quantum many-body problems: A Python implementation of Provably efficient machine learning for quantum many-body problems.
- The Annotated Transformer: An annotated version of the paper Attention Is All You Need by Vaswani et al., in the form of a line-by-line implementation.
Initial release (v1.0): Nov 2022
Current maintainers:
- Haoxiang Wang (@Haoxiang__Wang, hwang264@illinois.edu),
- Maurice Weber (@mauriceweberq, maurice.weber@inf.ethz.ch)
Apache 2.0