Morpho-MNIST: Quantitative Assessment and Diagnostics for Representation Learning – Now published in the Journal of Machine Learning Research!
Revealing latent structure in data is an active field of research, having introduced exciting technologies such as variational autoencoders and adversarial networks, and is essential to push machine learning towards unsupervised knowledge discovery. However, a major challenge is the lack of suitable benchmarks for an objective and quantitative evaluation of learned representations. To address this issue we introduce Morpho-MNIST, a framework that aims to answer: "to what extent has my model learned to represent specific factors of variation in the data?" We extend the popular MNIST dataset by adding a morphometric analysis enabling quantitative comparison of trained models, identification of the roles of latent variables, and characterisation of sample diversity. We further propose a set of quantifiable perturbations to assess the performance of unsupervised and supervised methods on challenging tasks such as outlier detection and domain adaptation.
If you use these tools or datasets in your publications, please consider citing the accompanying paper with a BibTeX entry similar to the following:
@article{castro2019morphomnist,
author = {Castro, Daniel C. and Tan, Jeremy and Kainz, Bernhard and Konukoglu, Ender and Glocker, Ben},
title = {{Morpho-MNIST}: Quantitative Assessment and Diagnostics for Representation Learning},
year = {2019},
journal = {Journal of Machine Learning Research},
volume = {20},
number = {178},
eprint = {arXiv:1809.10780}
}
Castro, D. C., Tan, J., Kainz, B., Konukoglu, E., & Glocker, B. (2019). Morpho-MNIST: Quantitative Assessment and Diagnostics for Representation Learning. Journal of Machine Learning Research, 20(178).
Table of Contents
We distribute the datasets in .zip files containing:
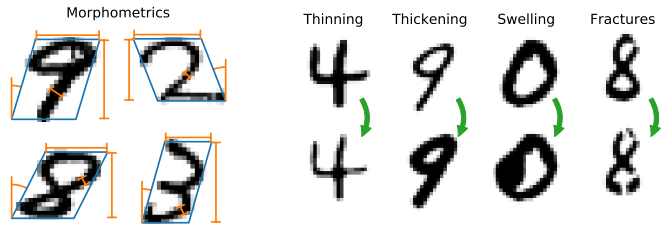
[train|t10k]-images-idx3-ubyte.gz: images[train|t10k]-labels-idx1-ubyte.gz: digit labels, copied from original MNIST[train|t10k]-morpho.csv: morphometrics table, with columns:index: index of the corresponding digit (for convenience, although rows are written in order)area(px²),length(px),thickness(px),slant(rad),width(px),height(px): calculated morphometrics
[train|t10k]-pert-idx1-ubyte.gz(globalandlocaldatasets): perturbation labels0: plain;1: thinned;2: thickened;3: swollen;4: fractured.
README-xxxx.txt: similar information to the above, for offline reference
Here are the downloads for the datasets used in the experiments in the paper:
| Dataset | Description | Download |
|---|---|---|
plain |
plain digits only | link (16 MB) |
global |
plain+thinning+thickening | link (15 MB) |
local |
plain+swelling+fractures | link (16 MB) |
We additionally provide the datasets affected by a single perturbation, from which local and global were composed by random interleaving with plain:
| Dataset | Description | Download |
|---|---|---|
thin |
thinning only | link (13 MB) |
thic |
thickening only | link (16 MB) |
swel |
swelling only | link (17 MB) |
frac |
fractures only | link (16 MB) |
Finally, we also make available the pre-computed morphometrics for the original MNIST images (only the .csv tables; the images and labels can be downloaded from LeCun's website):
| Dataset | Description | Download |
|---|---|---|
original |
original MNIST morphometrics | link (3.2 MB) |
The folder with all datasets for download can be accessed here.
The most relevant modules for end-users are io, measure and perturb, whose API we summarise below. For further details on these and on the other modules, please refer to the respective docstrings. The default arguments to all functions and constructors are the ones used in the paper and work well in practice.
Utility functions to load and save MNIST data files in IDX format. Can read and write plain or gzip-ed files, given the *.gz file extension.
input_images = load_idx("input_dir/images-idx3-ubyte.gz")
# ...
save_idx(output_images, "output_dir/images-idx3-ubyte.gz")Functions to compute morphometrics for a single MNIST image:
area, length, thickness, slant, width, height = measure_image(image)or for a batch of images, with support for parallel processing (can take up to a few minutes; displays a progress bar if tqdm is installed):
with multiprocessing.Pool() as pool:
metrics = measure_batch(images, pool=pool) # A pandas.DataFrameContains a number of subclasses of Perturbation, which apply a parametrisable transformation to a high-resolution binary MNIST image:
Perturbation(abstract)Thinning: Thin a digit by a specified proportion of its thickness.Thickening: Thicken a digit by a specified proportion of its thickness.Deformation(abstract)Swelling: Create a local swelling at a random location along the skeleton. Coordinates within a specified radius of the centre location are warped according to a radial power transform.
Fracture: Add fractures to a digit. Fractures are added at random locations along the skeleton, while avoiding stroke tips and forks, and are locally perpendicular to the pen stroke.
Perturbation instances are callable, taking as argument a morphomnist.morpho.ImageMorphology object constructed from the input image.
Below is a simple usage example applying a random perturbation (or none) to each of a collection of images:
import numpy as np
from morphomnist import io, morpho, perturb
perturbations = (
lambda m: m.binary_image, # No perturbation
perturb.Thinning(amount=.7),
perturb.Thickening(amount=1.),
perturb.Swelling(strength=3, radius=7),
perturb.Fracture(num_frac=3)
)
images = io.load_idx("input_dir/images-idx3-ubyte.gz")
perturbed_images = np.empty_like(images)
perturbation_labels = np.random.randint(len(perturbations), size=len(images))
for n in range(len(images)):
morphology = morpho.ImageMorphology(images[n], scale=4)
perturbation = perturbations[perturbation_labels[n]]
perturbed_hires_image = perturbation(morphology)
perturbed_images[n] = morphology.downscale(perturbed_hires_image)
io.save_idx(perturbed_images, "output_dir/images-idx3-ubyte.gz")
io.save_idx(perturbation_labels, "output_dir/pert-idx1-ubyte.gz")