This repository contains our tools & research for running DeepCell segmentation and QuPath measurements on Google Cloud Batch.
Our results show an overall improvement from ~13 hours to ~10 minutes for segmenting & measuring a cell. The starting point was running on a laptop or colo machine, and our work ran on GCP Batch with some cloud-focused enhancements.
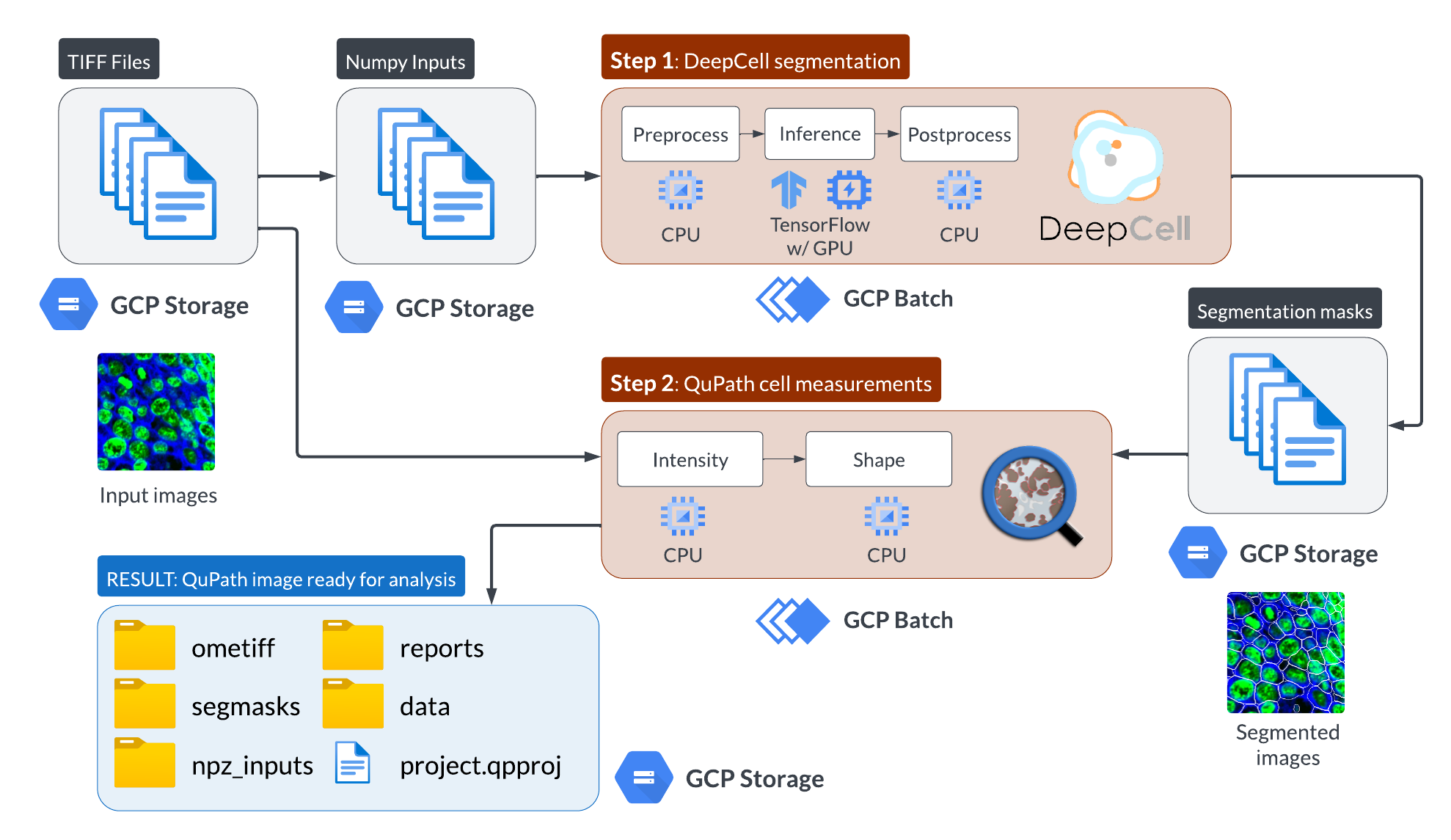
The workflow operates on one or more input images, converted from image to numpy pixel array. Then DeepCell preprocesses the data (denoising & normalization), runs the segmentation prediction, and postprocesses the predictions into a cell mask. Then, we load the image and mask into QuPath to compute quantitative metrics (size, channel intensity, etc.) for further analysis. For an example downstream usage, see SpaFlow (cell clustering & quantification).
Here is the workflow diagram:
You'll need a JSON file available in a cloud bucket, configuring the application environment. Create a file something like this:
{
"segment_container_image": "$REPOSITORY/benchmarking:latest",
"quantify_container_image": "$REPOSITORY/qupath-project-initializer:latest",
"bigquery_benchmarking_table": "$PROJECT.$DATASET.$TABLE",
"region": "$REGION",
"networking_interface": {
"network": "the_network",
"subnetwork": "the_subnetwork",
"no_external_ip_address": true
},
"service_account": {
"email": "the_service@account.com"
}
}You'll need to replace the variables with your environment.
- You can use the public Docker Hub containers, or copy them to your own artifact repository.
- For the benchmarking, you need to create a dataset & table in a GCP project; or you can omit it or set it to blank to skip collecting benchmarks. The table must be created with the schema specified in this file.
- Lastly, specify the GCP region where compute resources will be provisioned. This is not the same as storage buckets, but consider making it the same for efficiency & egress cost reduction.
- The
networking_interfaceandservice_accountsections are optional if you want to use default settings.
For example, using the Docker Hub containers & skipping benchmarking & default networking + service account:
{
"segment_container_image": "dchaley/deepcell-imaging:latest",
"quantify_container_image": "dchaley/qupath-project-initializer:latest",
"region": "us-central1"
}Upload this file somewhere to GCP storage. We put ours in the root of our working bucket. You'll pass this GS URI as a parameter to the scripts.
To run DeepCell on input images then compute QuPath measurements, use the helper scripts/segment-and-measure.py. There are two ways to run this script: (1) running on a QuPath workspace, and (2) running on explicit paths.
-
QuPath workspace:
-
Many QuPath projects are organized something like this:
📁 Dataset ↳ 📁 OMETIFF ↳ 🖼️ SomeTissueSample.ome.tiff ↳ 🖼️ AnotherTissueSample.ome.tiff ↳ 📁 NPZ_INTERMEDIATE ↳ 🔢 SomeTissueSample.npz ↳ 🔢 AnotherTissueSample.npz ↳ 📁 SEGMASK ↳ 🔢 SomeTissueSample_WholeCellMask.tiff ↳ 🔢 SomeTissueSample_NucleusMask.tiff ↳ 🔢 AnotherTissueSample_WholeCellMask.tiff ↳ 🔢 AnotherTissueSample_NucleusMask.tiff ↳ 📁 REPORTS ↳ 📄 SomeTissueSample_QUANT.tsv ↳ 📄 AnotherTissueSample_QUANT.tsv ↳ 📁 PROJ ↳ 📁 data ↳ ... ↳ 📄 project.qpprojTo generate segmentation masks & quantification reports, run the following command:
scripts/segment-and-measure.py --env_config_uri gs://bucket/path/to/env-config.json workspace gs://bucket/path/to/dataset
This will enumerate all files in the
OMETIFFdirectory that have matching files inNPZ_INTERMEDIATE, and run DeepCell segmentation to generate theSEGMASKnumpy files. Then it will run QuPath measurements to generate theREPORTSfiles.If your folder structure is different (for example
OME-TIFFinstead ofOMETIFF) you can use these parameters to specify the workspace subdirectories:--images_subdir,--npz_subdir,--segmasks_subdir,--project_subdir,--reports_subdir. Put these parameters after theworkspacecommand.
-
-
Explicit paths.
-
You can also specify all paths explicitly (the files don't have to be organized in a dataset). To do so, run this command:
scripts/segment-and-measure.py --env_config_uri gs://bucket/path/to/env-config.json paths --images_path gs://bucket/path/to/ometiffs --numpy_path gs://bucket/path/to/npzs --segmasks_path gs://bucket/path/to/segmasks --project_path gs://bucket/path/to/project --reports_path gs://bucket/path/to/reports
-
In either case, when you download the QuPath project, you'll need to download the OMETIFF files as well. When you open the project it will prompt you to select the base directory containing the OMETIFFs, and from there should automatically remap the image paths.
You can use the parameter --image_filter to only operate on a subset of the OMETIFFs. For example,
scripts/segment-and-measure.py
--env_config_uri gs://.../config.json
--image_filter SomeTissue
workspace gs://path/to/workspaceThis will operate on every file whose name begins with the string SomeTissue. This would match SomeTissueSample, SomeTissueImage, etc. Note that this parameter has to come before the workspace or paths parameter.
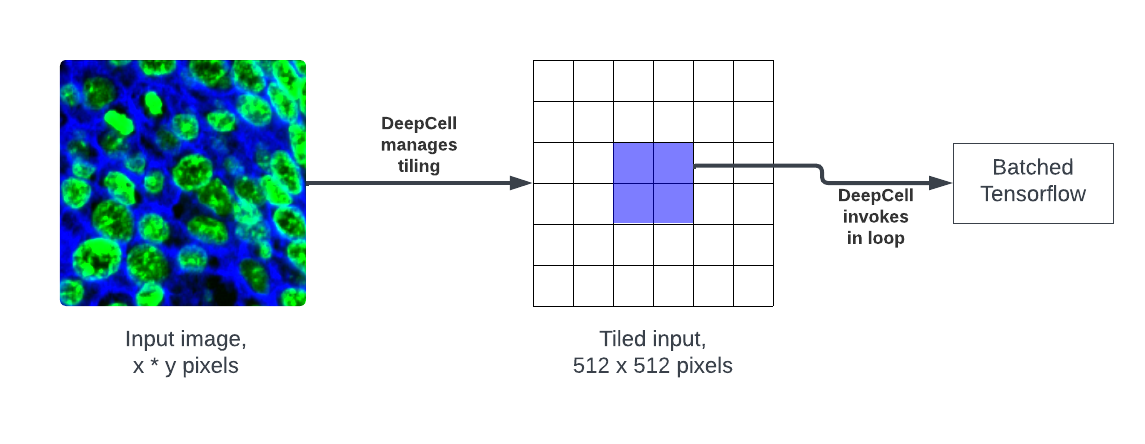
DeepCell does not process TIFF files. The TIFF channels must be extracted into Numpy arrays first.
DeepCell divides the preprocessed input into 512x512 tiles which it predicts in batches, then recombines into a single image for postprocessing.
This makes the prediction very resource-efficient, note however that pre- and post-processing still operate on the entire image. This is particularly problematic for post-processing which is very resource-intensive.
The prediction step outputs which pixels are most likely to be the center of their cell. The post-processing step runs image analysis algorithms to create the final cell masks. It operates a bit like a "flood fill" to expand the center out.
This uses the h_maxima grayscale reconstruction algorithm, which is (counterintuitively) far slower than prediction itself for large images.
Once we have cell predictions, we need to generate quantified metrics for the cells: location, size, channel intensities, and so on. This is crucial for downstream processing & analysis, including in a QuPath desktop environment. For example, a researcher might provide an analyzed & packaged QuPath project to a principal investigator for review.
QuPath is distributed as JAR files. Bioinformaticians typically run Groovy scripts in the embedded QuPath environment, however we don't have a desktop or VM environment for that. Instead we compile Kotlin code with the JARs to run on Google Batch.
The source code for quantifying the metrics plus building the container is located in a different repository: qupath-project-initializer.
QuPath measurements are computed a cell at a time. The algorithm re-fetches the image region containing the cell for each cell. This is prohibitively expensive for bulk measurement.
Adding code to prefetch the image into memory, then retrieve subregions from memory, provided a dramatic ~99% speed-up.
- GOAL: Understand and optimize DeepCell cellular segmentation on GCP at scale.
- KEY LINK #1: our benchmarking process.
- KEY LINK #2: our support/testing notebooks.
- KEY LINK #3: our project board & work areas for this project.
GPU makes a dramatic difference in model inference time.
Memory usage increases linearly with number of pixels.
Here are some areas we've identified:
- Preprocessing
- DeepCell converts everything to 64bit float. That's memory intensive. Do we actually need to?
- Postprocessing
- h_maxima: need to ship a ~15x speedup optimization
- Cost
- Run the prediction phase only with GPU infrastructure. Run everything else with CPU-only infrastructure.
This repo uses git-lfs (Git Large File System) to exclude large files (like sample numpy data) in the source history. This process is automatic & transparent, but requires git-lfs to be installed beforehand. Please see these instructions.
TLDR,
- on Mac,
brew install git-lfs. - on Linux,
sudo [apt-get | yum] install git-lfs. - on Windows,
git-lfsis included in the Git distribution.
Nothing special. You just need Python 3.10 at the latest.
python3.10 -m venv venv
source venv/bin/activate
pip install -r requirements.txt
Some incantations are needed to work on Apple silicon computers. You also need Python 3.9.
DeepCell depends on tensorflow, not tensorflow-macos. Unfortunately we need tensorflow-macos specifically to provide TF2.8 on arm64 chips.
The solution is to install the packages one at a time so that the DeepCell failure doesn't impact the other packages.
python3.9 -m venv venv
source venv/bin/activate
pip install -r requirements-mac-arm64.txt
cat requirements.txt | xargs -n 1 pip install
# Let it fail to install DeepCell, then:
pip install -r requirements.txt --no-deps
# Lastly install our own library. Note --no-deps
pip install --editable . --no-deps
I think but am not sure that the first --no-deps invocation is unnecessary as pip install installs dependencies.