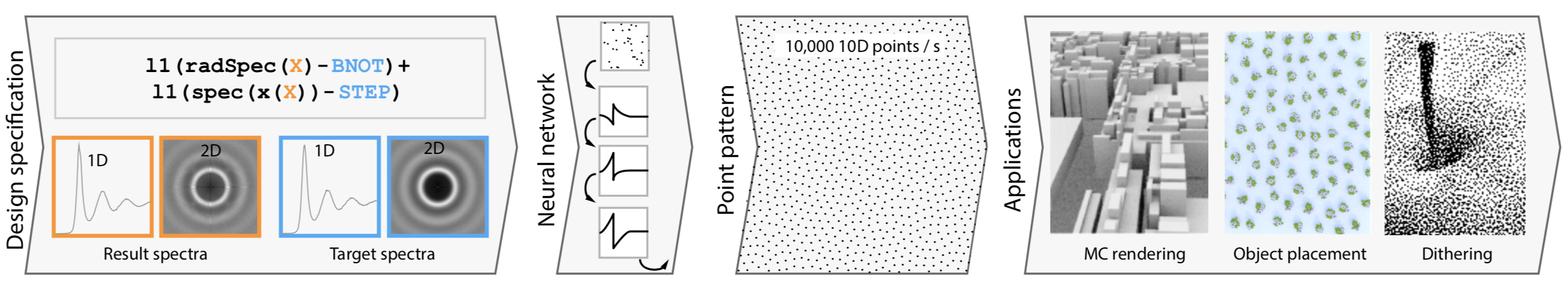
Authors: Thomas Leimkühler, Gurprit Singh, Karol Myszkowski, Hans-Peter Seidel, Tobias Ritschel.
The paper and supplemental material can be found on our project webpage.
@article{leimkuehler2019sigasia,
title={Deep Point Correlation Design},
author={Thomas Leimk\"uhler and Gurprit Singh and Karol Myszkowski and Hans-Peter Seidel and Tobias Ritschel},
journal={ACM Transactions on Graphics (Proceedings of SIGGRAPH Asia 2019)},
volume={38},
number={6}
year={2019},
doi={10.1145/3355089.3356562}
}
To check out the code on a local machine run
git clone https://github.com/sinbag/deepsampling.git
This code was developed using Tensorflow v1.9 with Python v3.6 and relies on the following packages:
matplotlib, imageio, openEXR
To compile the custom tensorflow filtering op, you can use cmake by adapting the paths INCLUDE_DIRECTORIES() and SET(tensorflowLib "") in
/ops/cmake/CMakeLists.txt
Execute the following commands on the terminal to compile and build the cmake file:
cd deepsampling/ops/
cmake/createBuild.sh
cmake --build ./build
After compilation, adapt CUSTOM_OP_PATH in
/ops/src/filter/dist_filter_gpu.py
to point to the libCustomOp.so file you created.
We are ready to run our first experiment. You can start with the example files provided in the src/experiments/folder. Go to the folder containing the main.py file and execute the following command to start training for Fig.8a from the paper.
python main.py -e experiment_fig8a
Note that the .py extension of the experiment file is not provided. To run any other experiment file, simply run
python main.py -e <your_experiment_filename>
where <your_experiment_filename> refers to the name of the file you can create following the instructions below.
To define a network and a design specification, you need to set up a Python experiment file and put it into
src/experiments/
You will find pre-defined experiment files in this directory, so you can just start from these. The pre-defined experiments correspond to figures in the paper and are meant for easy reproduction of results.
First, you need to define three global variables: LOGS_DIR is the output directory, where network outputs (spectra, realizations, ...) and trained weights are written to. TARGET_DIR points to the directory where target spectra and realizations are stored. FILE_EXT is the preferred visualization output format (usually ".pdf" or ".png").
Next, you need to implement the buildEnvironment() function.
Specify a TrainingSetup with the following parameters:
| Variable | Meaning | Symbol in paper |
|---|---|---|
pointCount |
Number of points. | m |
dimCount |
Number of dimensions. | n |
batchSize |
Number of point sets in a batch. Used for variance reduction. | - |
griddingDims |
Number of gridded dimensions. Set this to 0 if you don't want to use gridding. |
- |
convCount |
Length of the filter sequence. | l |
kernelCount |
Number of unique filter kernels used. These kernels are distributed over the filters by linear interpolation of weights. | l_u |
kernelSampleCount |
Resolution of the filter kernels. | b |
receptiveField |
Receptive field of the filter kernels. | r |
projectionsStrings |
Here you specify which (sub-)spaces you want the filters to operate in by using a list of strings. Digits correspond to dimensions. Example 1 (full space): You have set dimCount = 2. Then ['01'] lets the filters operate in full two-dimensional space. Example 2 (projective subspaces): You have set dimCount = 3. Then ['012','01','12','02'] lets the filters operate in full three-dimensional space and in all canonical 2D subspaces. |
M |
customOp |
Set to true if you want to use the custom filter implementation (recommended). |
- |
trainIterations |
Training iterations. | - |
learningRate |
Learning rate. | - |
displayGrid |
Set to true if you want to display a grid on top of point visualizations. |
- |
evalRealizations |
Number of point set realizations to estimate expected power spectra or differential histograms outside of the training loop. This number also specifies the number of realizations to export if saveEvalRealizations is set to true. |
- |
saveEvalRealizations |
Set to true if you want to export network output point sets (the number is determined by evalRealizations). |
- |
storeNetwork |
Set to true if you want to store trained network weights. This happens at the end of training and in regular intervals, as specified in backupInterval. |
- |
backupInterval |
The frequency (in training iterations) at which the network weights will be stored during training. | - |
weightDir |
The directory to load network weights from. If you want to train from scratch, set this to None. |
- |
If you want to use the code in deployment mode, set trainIterations = 0 and load your network weights by specifying weightDir.
Now you need to specify any targets that you want to use. You can define any number of spectra and differential histograms by concatenating them into lists. Keep two lists: One for spectra, one for histograms.
To set up a spectrum target, create a FourierSetup with the following parameters:
| Variable | Meaning | Symbol in paper |
|---|---|---|
resolution |
Resolution of the spectrum along one axis. | n_c |
cancelDC |
Set to true if you want to ignore the DC term (recommended). |
- |
mcSamplesPerShell |
Number of samples per radial frequency when using a Monte Carlo estimation of a radial spectrum. | - |
Now you can load or estimate any spectrum using the methods provided in the FourierSetup class in src/setup.py, e.g., load a 2D spectrum from an EXR file using loadTarget2D().
To set up a histogram target, create a HistogramSetup with the following parameters:
| Variable | Meaning | Symbol in paper |
|---|---|---|
trainingSetup |
The training setup you created ealier. | - |
binCount |
Resolution of the histogram. | n_c |
stdDev |
The standard deviation of the Gaussian weights. | - |
scale |
Scaling of distances to focus on the relevant range. | - |
mcSamples |
Number of point set realizations used to estimate the histogram during setup. | - |
Now you can estimate any differential histogram using the methods provided in the HistogramSetup class in src/setup.py, e.g., estimate a histogram from realizations using createTargetFromData().
Once you have created a training setup and a list of Fourier and/or differential histogram setups, wrap them into an environment and return it:
return setup.Environment(trainingSetup, fourierSetupList, histogramSetupList)
If you don't use a histogram or a Fourier setup, just put None.
Lastly, you need to specify the loss function by implementing lossSetup(env, outputNode). Through the environment env you have access to all information and targets you created earlier. The outputNode simply is the output of the last filtering operation of the network, i.e., the resulting point set(s).
You need to return the loss value and, optionally, (lists of) spectra and histograms you want to visualize. A simple example of a loss implementation which L1-compares the Monte Carlo estimate of the radially averaged spectrum of the network's output to a reference looks like this:
spectrumNode = fourier.radialSpectrumMC(outputNode, env.fourierSetupList[0])
lossNode = l1Loss(spectrumNode, env.fourierSetupList[0].target)
histogramNode = None
return lossNode, spectrumNode, histogramNode