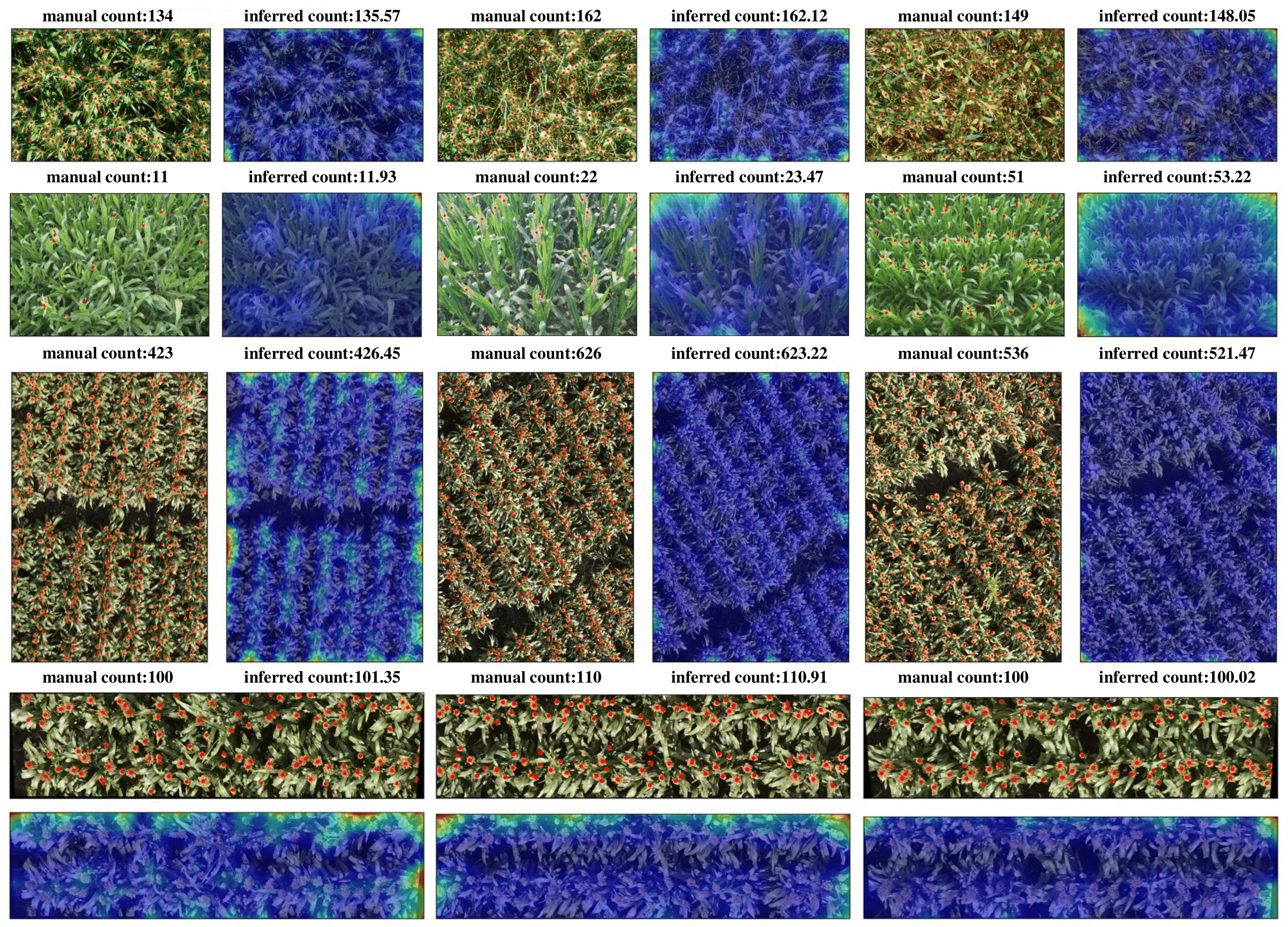
This repository includes the official implementation of TasselNetv2+ for plant counting, presented in paper:
TasselNetv2+: A Fast Implementation for High-Throughput Plant Counting from High-Resolution RGB Imagery
Frontiers in Plant Science, 2020
Hao Lu and Zhiguo Cao
- Highly Efficient: TasselNetv2+ runs an order of magnitude faster than TasselNetv2 with around 30fps on image resolution of 1980×1080 on a single GTX 1070;
- Effective: It retrains the same level of counting accuracy compared to its counterpart TasselNetv2;
- Easy to Use: Pretrained plant counting models are included in this repository.
The code has been tested on Python 3.7.4 and PyTorch 1.2.0. Please follow the official instructions to configure your environment. See other required packages in requirements.txt.
Wheat Ears Counting
- Download the Wheat Ears Counting (WEC) dataset from: Google Drive (2.5 GB). I have reorganized the data, the credit of this dataset belongs to this repository.
- Unzip the dataset and move it into the
./datafolder, the path structure should look like this:
$./data/wheat_ears_counting_dataset
├──── train
│ ├──── images
│ └──── labels
├──── val
│ ├──── images
│ └──── labels
Maize Tassels Counting
- Download the Maize Tassels Counting (MTC) dataset from: Google Drive (1.8 GB)
- Unzip the dataset and move it into the
./datafolder, the path structure should look like this:
$./data/maize_counting_dataset
├──── trainval
│ ├──── images
│ └──── labels
├──── test
│ ├──── images
│ └──── labels
Sorghum Heads Counting
- Download the Sorghum Heads Counting (SHC) dataset from: Google Drive (152 MB). The credit of this dataset belongs to this repository. I only use the two subsets that have dotted annotations available.
- Unzip the dataset and move it into the
./datafolder, the path structure should look like this:
$./data/sorghum_head_counting_dataset
├──── original
│ ├──── dataset1
│ └──── dataset2
├──── labeled
│ ├──── dataset1
│ └──── dataset2
Run the following command to reproduce our results of TasselNetv2+ on the WEC/MTC/SHC dataset:
sh config/hl_wec_eval.sh
sh config/hl_mtc_eval.sh
sh config/hl_shc_eval.sh
- Results are saved in the path
./results/$dataset/$exp/$epoch.
Run the following command to train TasselNetv2+ on the on the WEC/MTC/SHC dataset:
sh config/hl_wec_train.sh
sh config/hl_mtc_train.sh
sh config/hl_shc_train.sh
To use this framework on your own dataset, you may need to:
- Annotate your data with dotted annotations. I recommend the VGG Image Annotator;
- Generate train/validation list following the example in
gen_trainval_list.py; - Write your dataloader following example codes in
hldataset.py; - Compute the mean and standard deviation of RGB on the training set;
- Create a new entry in the
dataset_listinhltrainval.py; - Create a new
your_dataset.shfollowing examples in./configand modify the hyper-parameters (e.g., batch size, crop size) if applicable. - Train and test your model. Happy playing:)
If you find this work or code useful for your research, please cite:
@article{lu2020tasselnetv2plus,
title={TasselNetV2+: A fast implementation for high-throughput plant counting from high-resolution RGB imagery},
author={Lu, Hao and Cao, Zhiguo},
journal={Frontiers in Plant Science},
year={2020}
}
@article{xiong2019tasselnetv2,
title={TasselNetv2: in-field counting of wheat spikes with context-augmented local regression networks},
author={Xiong, Haipeng and Cao, Zhiguo and Lu, Hao and Madec, Simon and Liu, Liang and Shen, Chunhua},
journal={Plant Methods},
volume={15},
number={1},
pages={150},
year={2019},
publisher={Springer}
}
This code is only for non-commercial purposes. Please contact Hao Lu (hlu@hust.edu.cn) if you are interested in commerial use.