This repository hosts the R codebase for our data analysis and modelling workflows. Most of our scripts (*.R) include some roxygen2-style commenting for generating markdown report files (*.knit.md), shared here to better document and organize our research.
R/get_data contains R scripts to fetch public datasets on the hydrology of UYR:
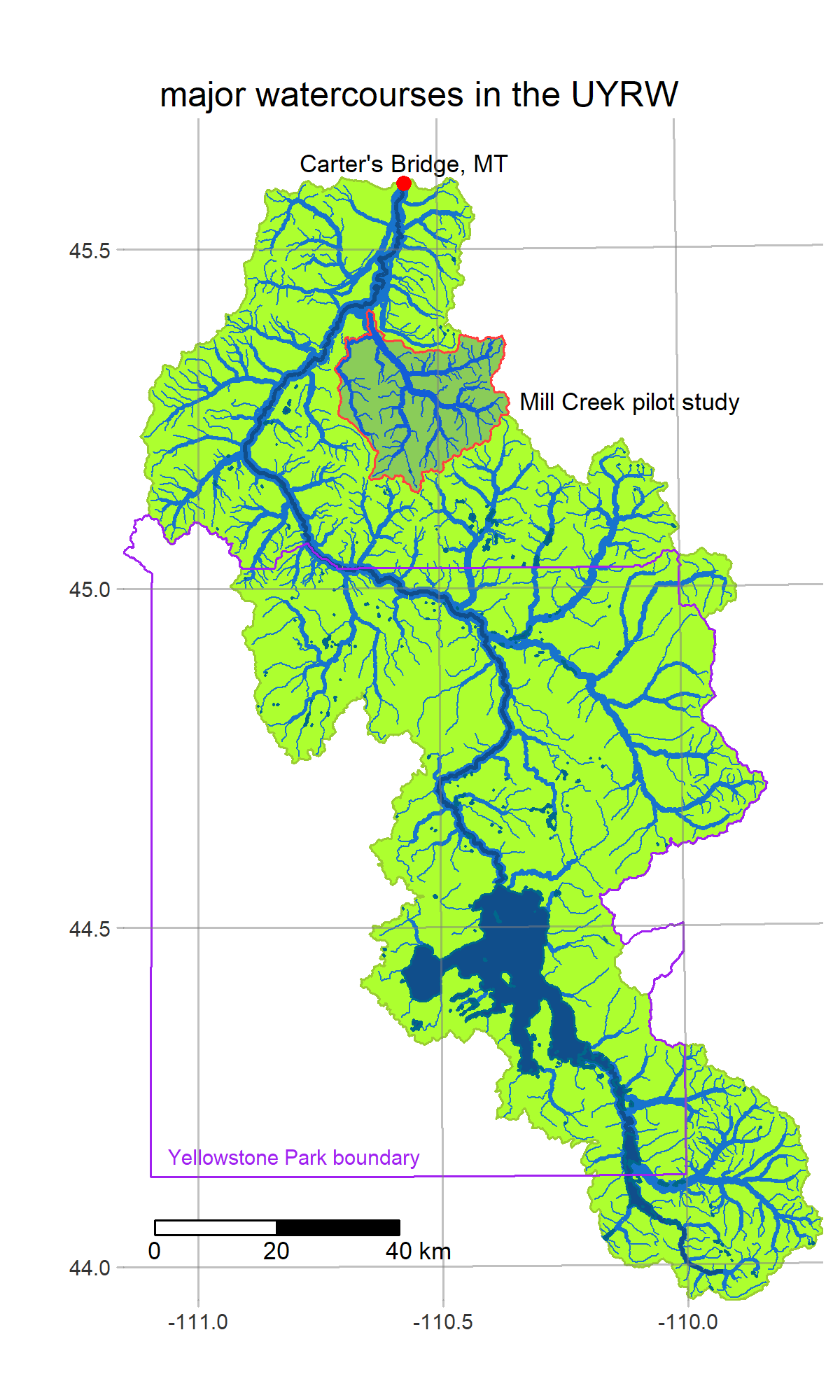
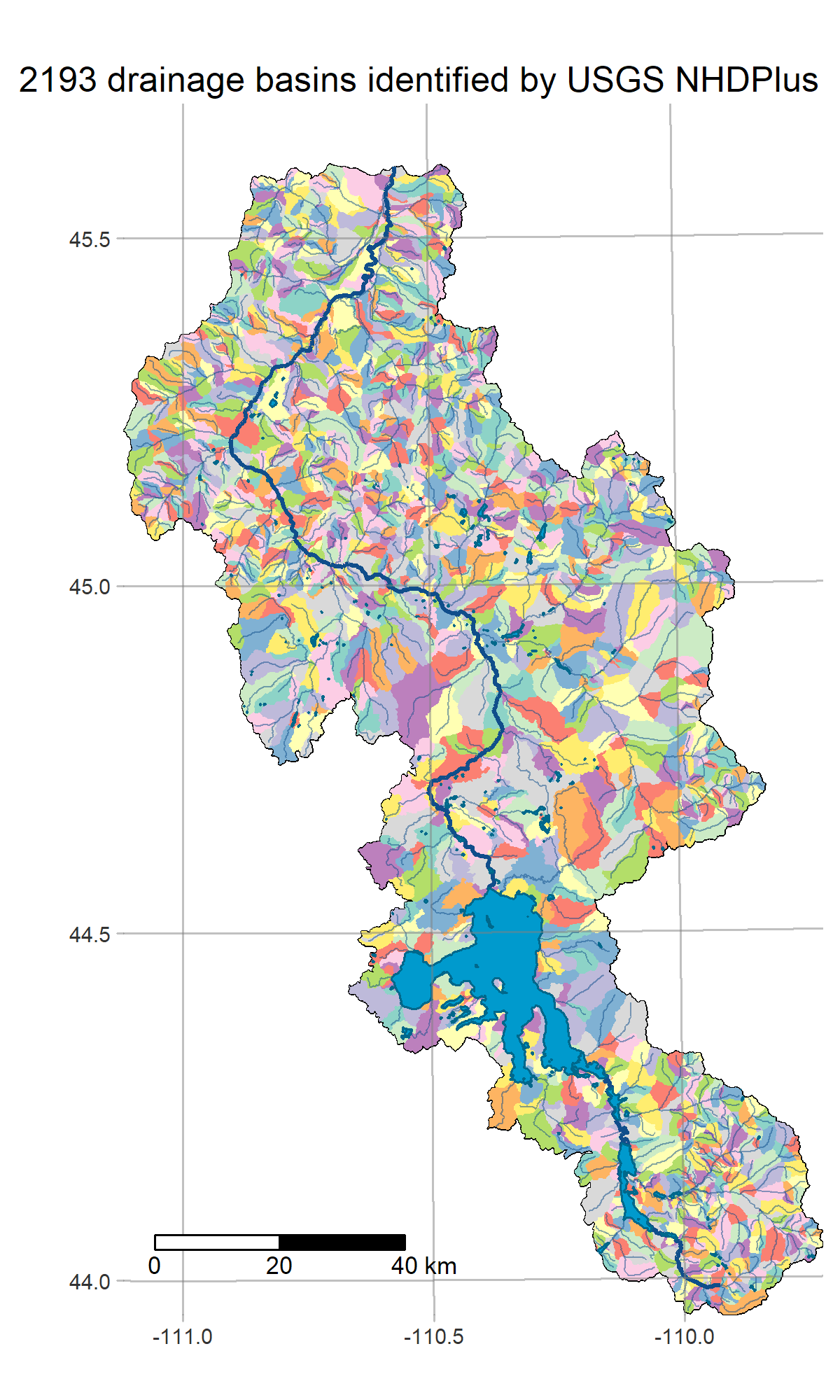
- get_basins defines the study area and loads some hydrology info from NHDPlus
- get_weatherstations finds climatic sensor station data at the GHCN
- get_dem fetches the USGS National Elevation Dataset
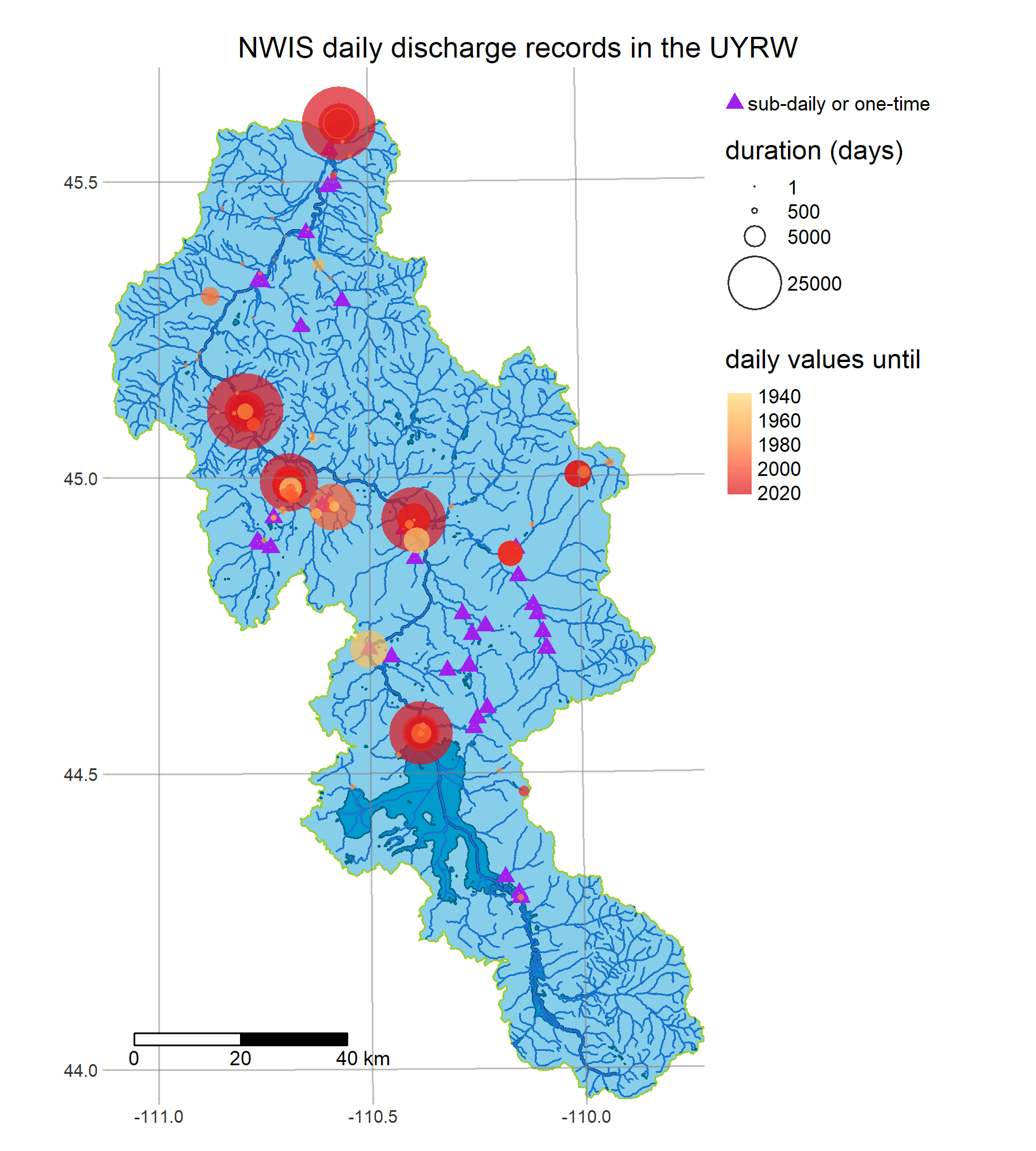
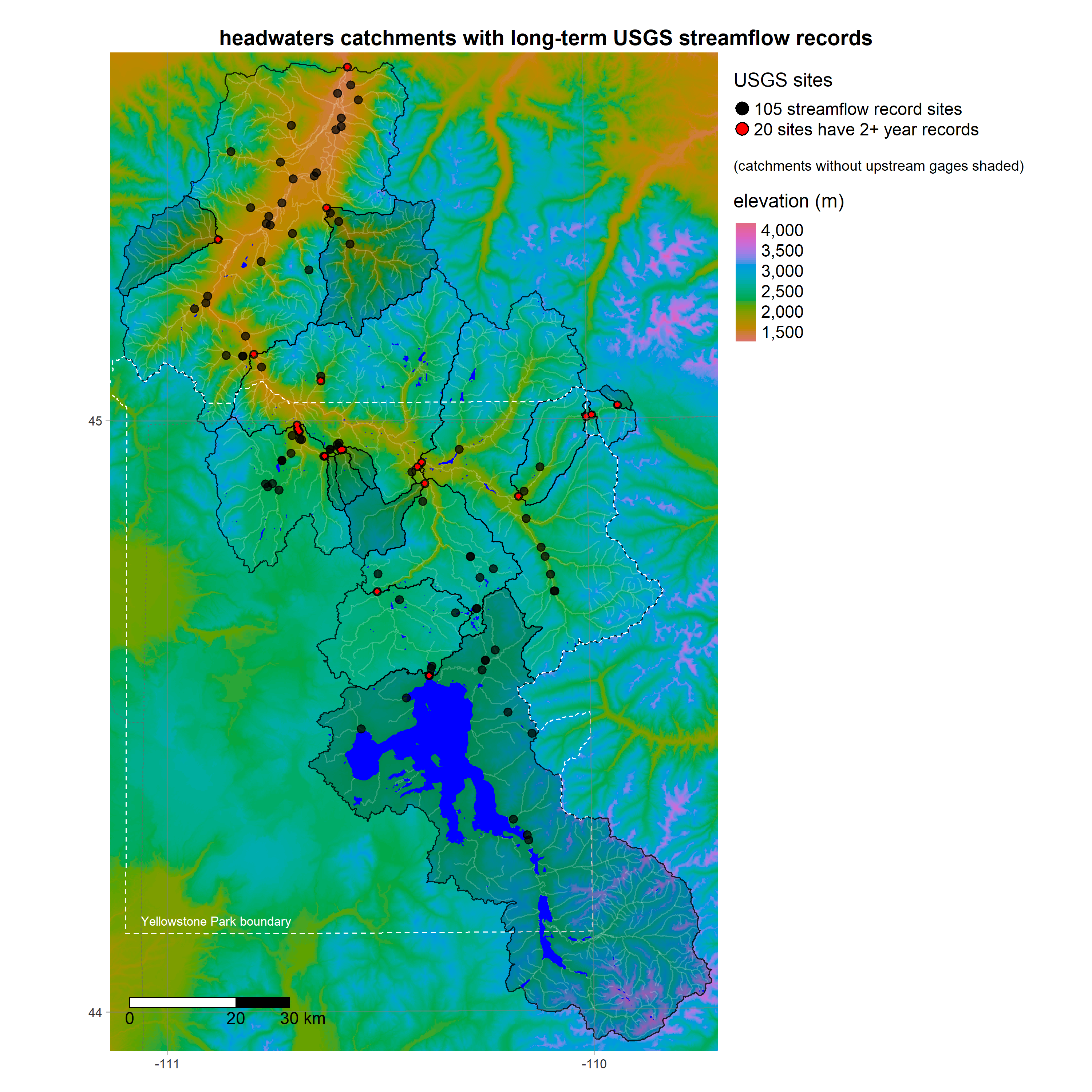
- get_streamgages finds historical daily values from USGS Water Services
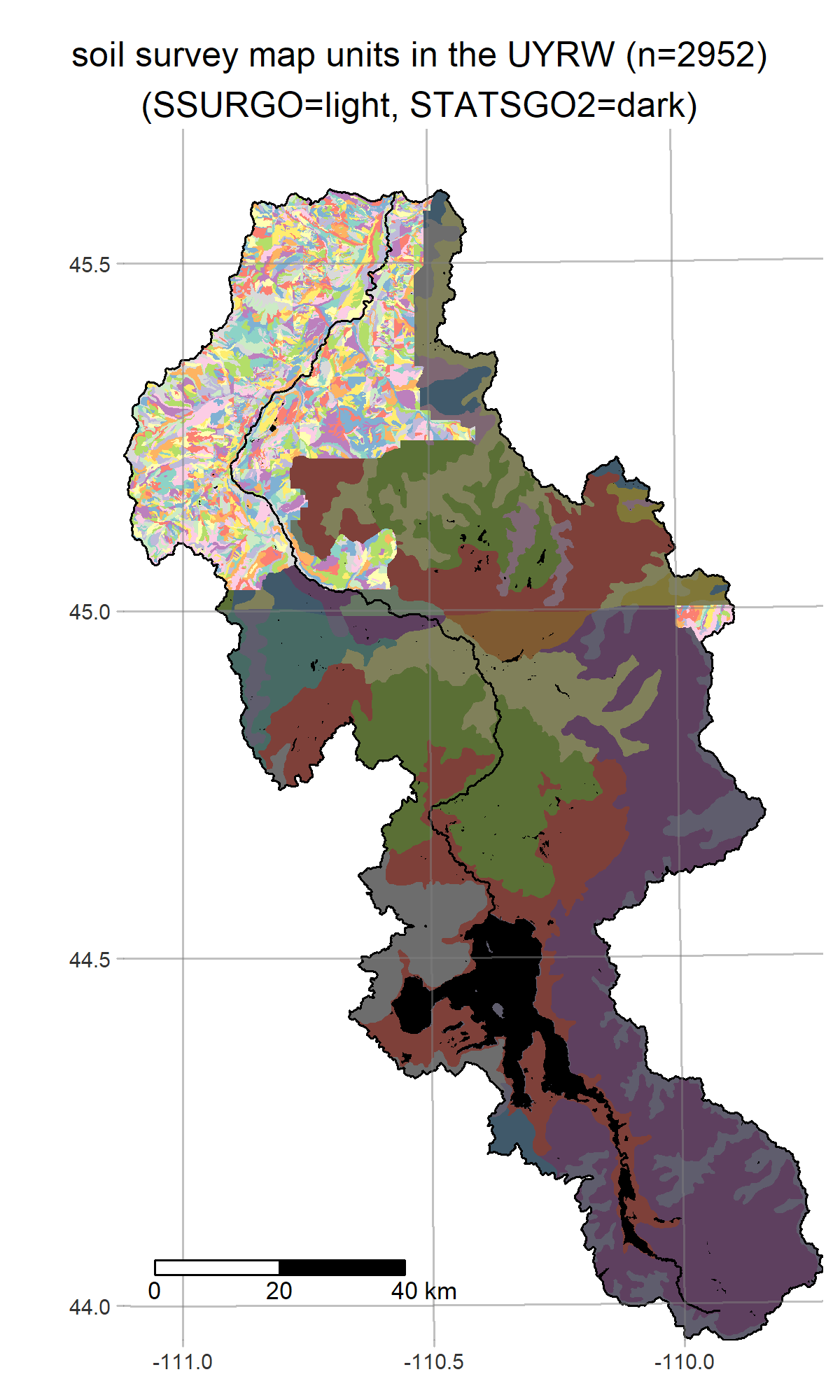
- get_soils fetches SSURGO/STATSGO2 maps from the National Cooperative Soil Survey
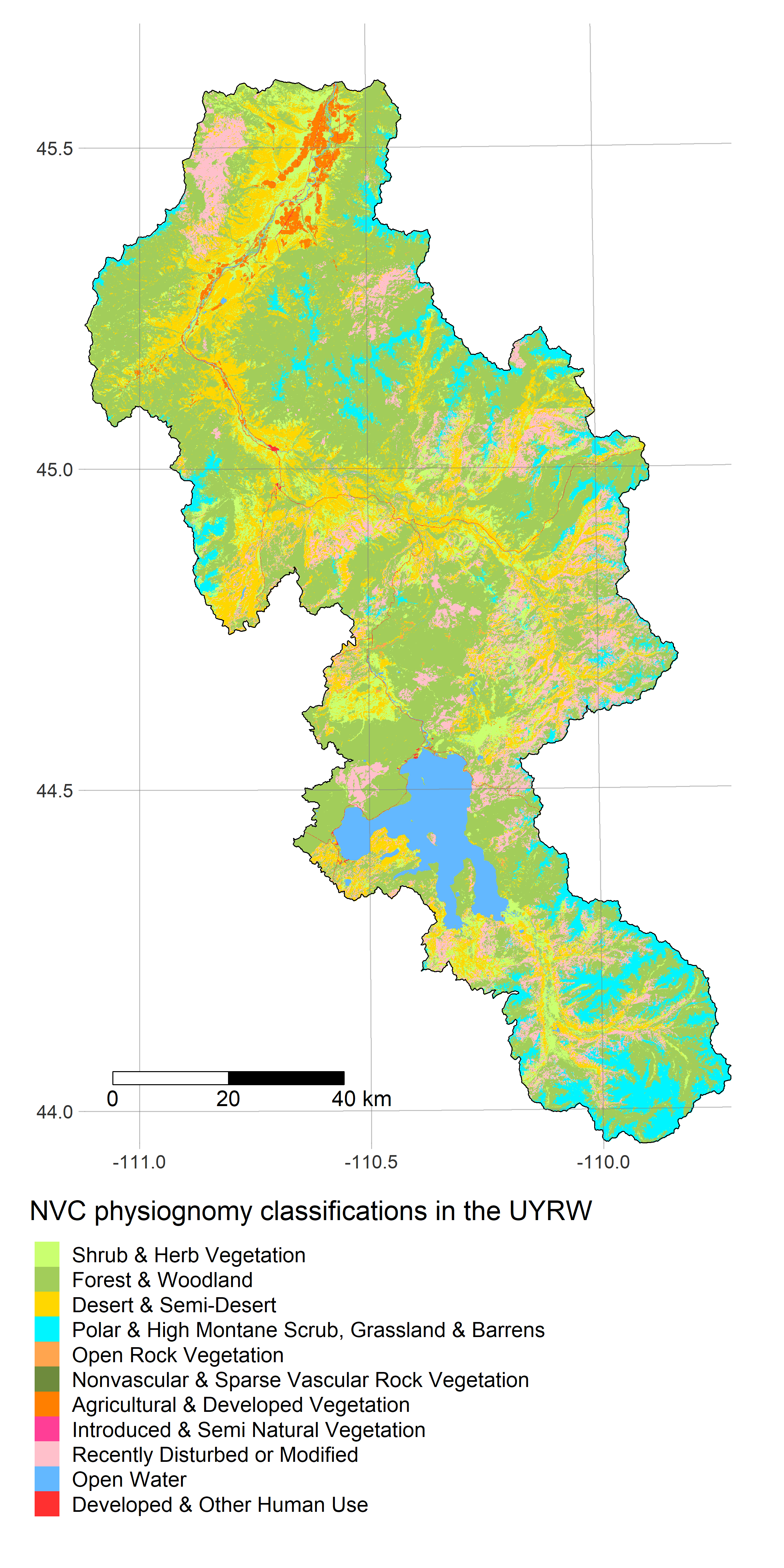
- get_landuse fetches data from GAP/LANDFIRE
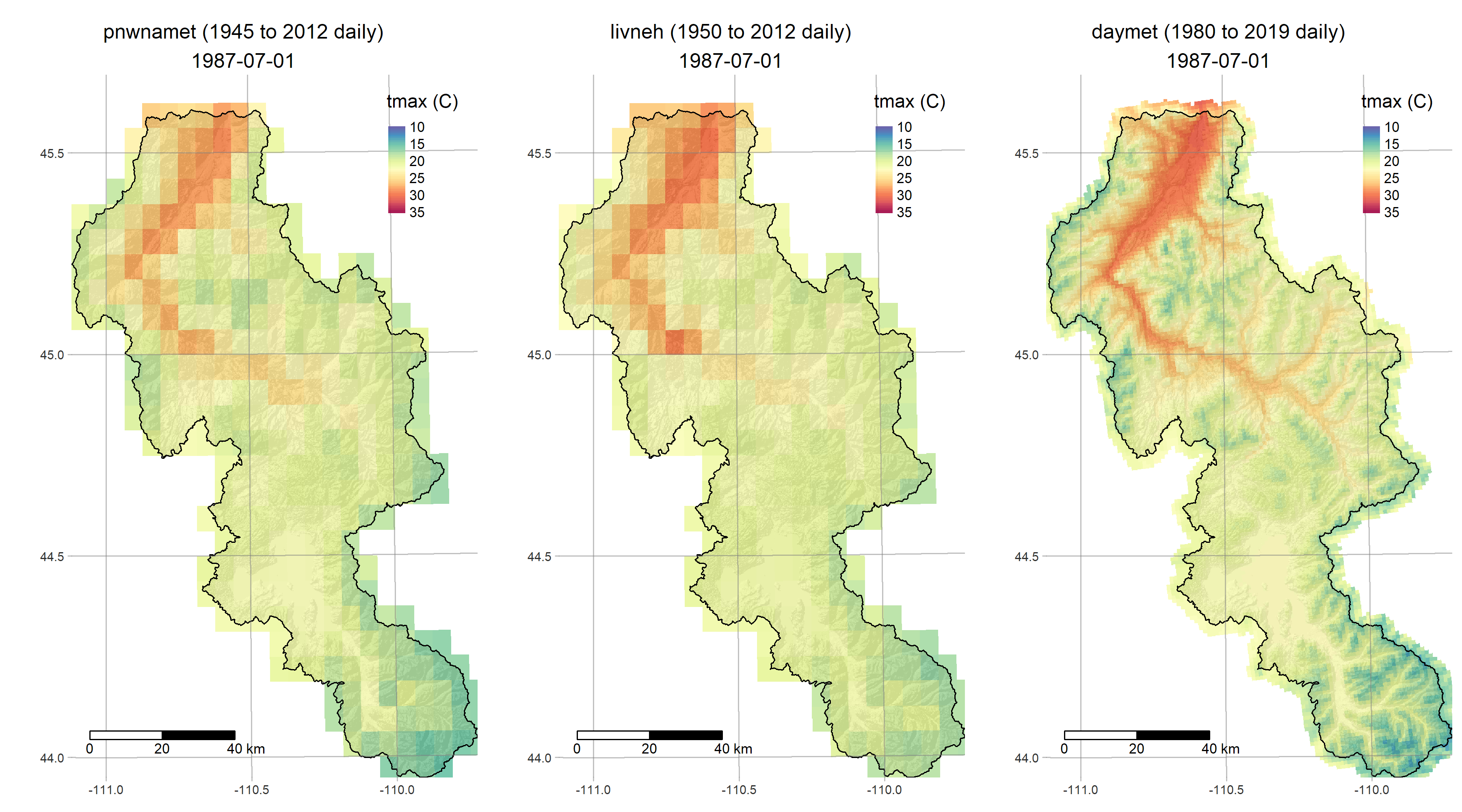
- get_meteo fetches three different gridded meteorological reconstructions
- get_snow finds snow data at SNOTEL and partner networks
The Soil and Water Assessment Tool (SWAT) is a widely-used river basin watershed model with a very long development history dating back to the early 90s. Read more about it here. These days SWAT has a large and active community of users running one of the two public-domain Fortran-based implementations maintained by the USDA and Texas A&M:
-
SWAT2012 is the legacy version. Advantages include: a larger community, a very long development and testing history, more extensions and helper software available, better documentation, and a larger presence in the hydrology literature.
-
SWAT+ is the newer revised version. Advantages include more flexibility in spatial representation of watershed features, a modernized file structure. This seems the likely focus of future R&D by the SWAT development team.
Both versions continue to be updated and patched regularly (don't be fooled by the "2012"). However, although the mathematical models behind the names are largely the same, their project files are not interchangeable as SWAT+ uses different variable names and file structures. Our project will focus on building extensions of the newest official release of SWAT+.
The R/analysis directory (in development) is for building, fitting and analysing SWAT+ models in R.
Using the output from /R/get_data, we plan to fit SWAT+ models sequentially on small catchments in the UYRW. Check back in the coming weeks as we add to this section to demonstrate the model building code:
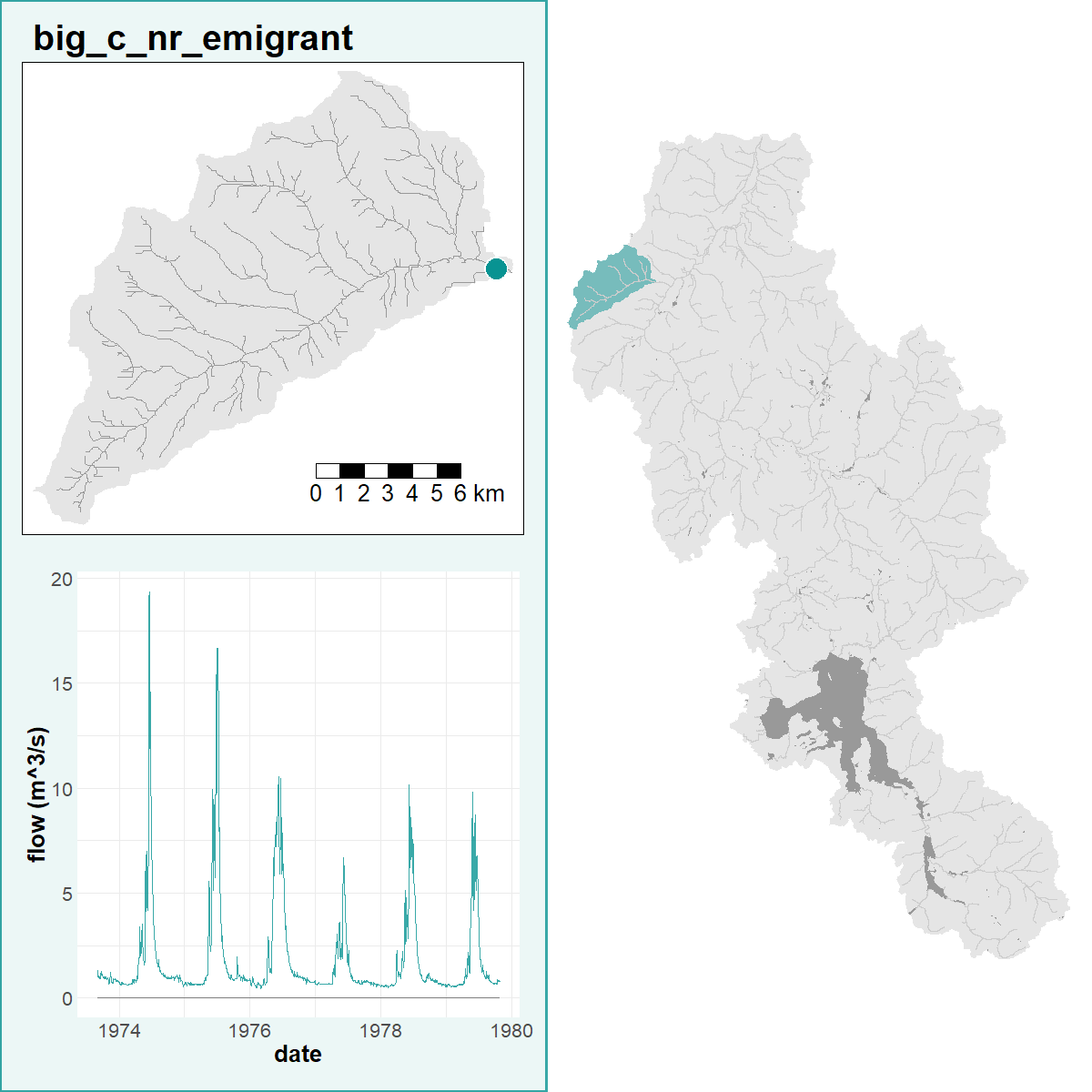
- make_subwatersheds partitions the UYRW area into subwatersheds with USGS gages at their outlets
- helper_analysis utilities for creating and calibrating SWAT+ models in R
The parametrization of a SWAT model is not trivial. A typical use case will involve dozens of config files containing many thousands of model parameters. Fortunately, SWAT is largely made up of process-based components whose physically-based parameters can (at least initially) be set using empirical data.
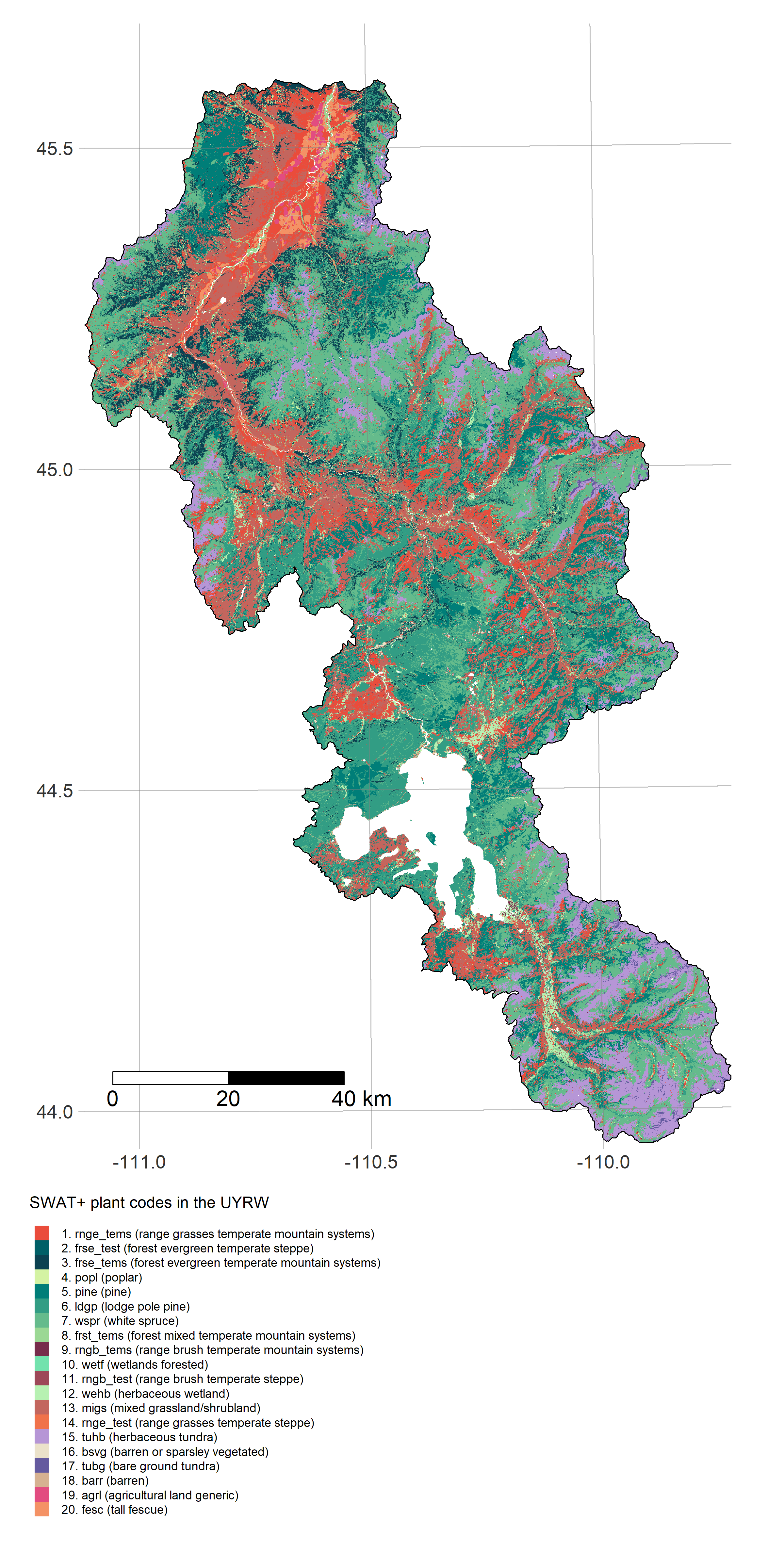
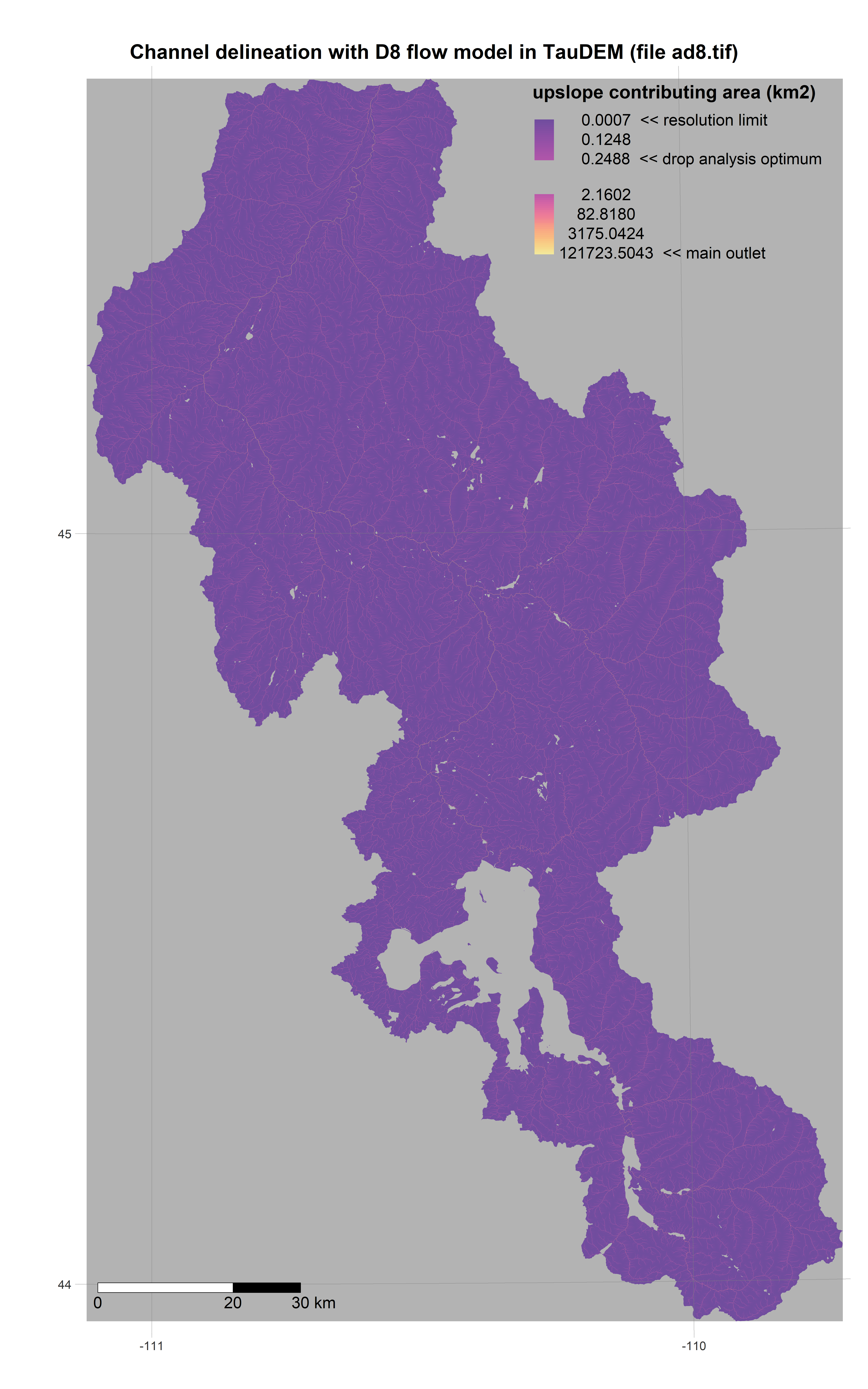
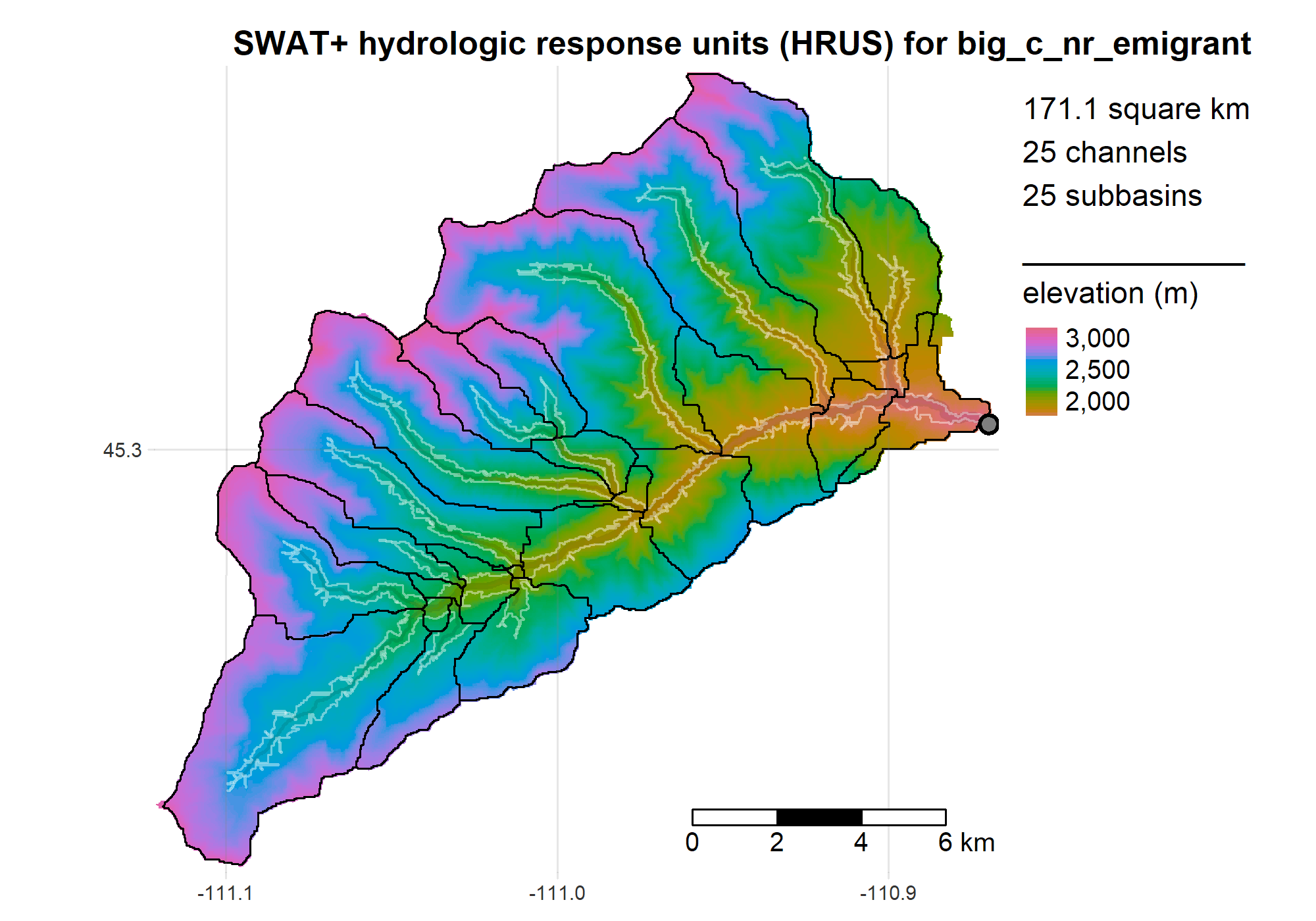
For example after running get_dem to fetch our DEM, we can run the TauDEM workflow to define subbasins and construct a routing network connecting them. After running get_soils and get_landuse, we can then delineate HRUs and assign their plant/soil parameters to survey values. These steps are usually carried out using QSWAT/QSWAT+ (GUI-based QGIS plugins). Our code runs them programmatically from within R:
rswat is a set of tools for building SWAT+ models and managing their many files. This includes R wrappers for running the QSWAT+ workflow in PyQGIS, calling SWATEditor.exe to build model config files; reading, cataloging and modifying these files, executing simulations, and reading the outputs. The following vignettes demonstrate these features on a simple example catchment
- demo_qswat builds the SWAT+ model
- demo_txtinout loads and modifies it
- demo_objective tunes parameters
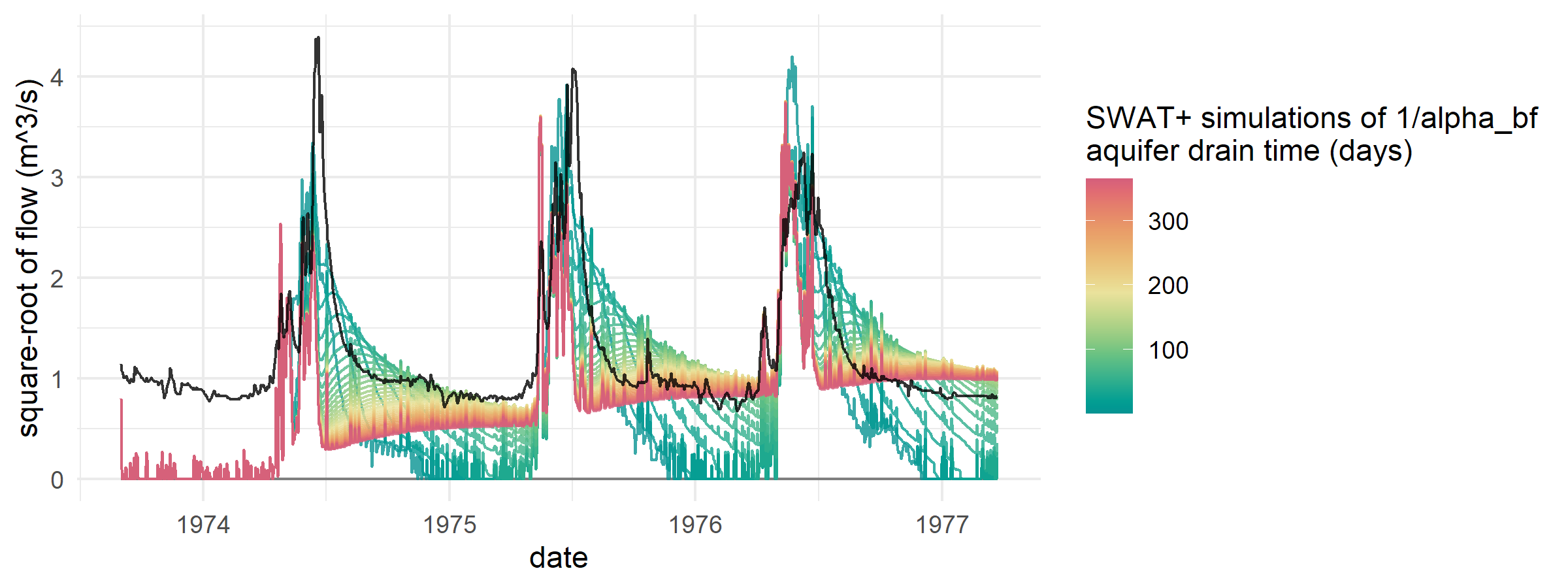
- demo_baseflow adjusts aquifer parameters
- swat_outputs_API exports model outputs
rswat_docs is a small set of R functions that renders the inputs and outputs documentation PDFs as text and extracts a large, easily searchable table of name-definition pairs. See demo_rswat_docs for a vignette on searching the I/O PDF from within R.
We may polish some/all of this code and release it as an R package at some point. Those interested in script-based workflows for SWAT are encouraged to also check out the SWAT+AW and SWATPlusR projects.
We are a team of mathematicians, statisticians, and ecologists, conducting a multi-year research project to develop an operational forecasting system for streamflow and water quality on the Upper Yellowstone River (UYR) and its tributaries. Our system will be based on SWAT+/gflow, a hybrid of the SWAT+ (Soil-Water-Assessment Tool) model for surface water dynamics, and gflow, physically-based groundwater flow module developed by Ryan Bailey to replace the current SWAT+ aquifer module.
Our work is funded through a MITACS Accelerate International grant to Dean Koch, partnering the University of Alberta with R2CS LLC in Montana, and the Yellowstone Ecological Research Center. The project began on August 3, 2020.
R is a powerful data-retrieval, GIS, and visualization tool. These figures are generated by the scripts in our repo using the tmap package: