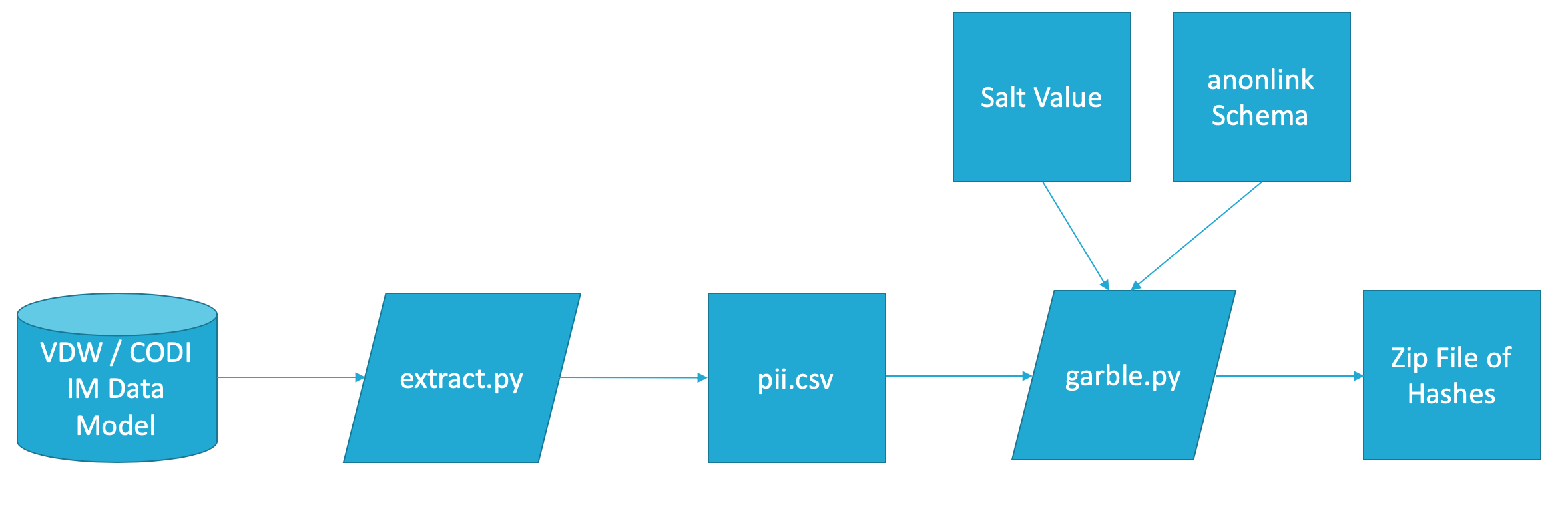
Tools for Childhood Obesity Data Initiative (CODI) data owners to extract personally identifiable information (PII) from the CODI Data Model and garble PII to send to the data coordinating center (DCC) for matching. These tools facilitate hashing / Bloom filter creation part of a Privacy-Preserving Record Linkage (PPRL) process.
- Installation
- Overall Process
- Extract PII
- Garbling
- Mapping LINKIDs to PATIDs
- Additional Information for Developer Testing and Tuning
- Notice
These tools were created and tested on Python 3.7.4. The tools rely on two libraries: SQLAlchemy and clkhash.
SQLAlchemy is a library that allows the tools to connect to a database in a vendor independent fashion. This allows the tools to connect to a database that conforms to the CODI Identity Data Model implented in PostgreSQL or Microsoft SQLServer (and a number of others).
clkhash is a part of the anonlink suite of tools. It is repsonsible for garbling the PII so that it can be de-identified prior to transmission to the DCC. Note: you may have to specify the latest anonlink docker image when pulling, to ensure you are on the right version as registry may have old version (tested on v1.14)
-
Download the tools as a zip file using the "Clone or download" button on GitHub.
-
Unzip the file.
-
From the unzipped directory run:
pip install -r requirements.txt
N.B. If the install fails during install of psycopg2 due to a clang error, you may need to run the following to resolve:
env LDFLAGS='-L/usr/local/lib -L/usr/local/opt/openssl/lib -L/usr/local/opt/readline/lib' pip install psycopg2==2.8.4
- Install Anaconda by following the install instructions.
- Depending on user account permissions, Anaconda may not install the latest version or may not be available to all users. If that is the case, try running
conda update -n base -c defaults conda
- Depending on user account permissions, Anaconda may not install the latest version or may not be available to all users. If that is the case, try running
- Download the tools as a zip file using the "Clone or download" button on GitHub.
- Unzip the file.
- Open an Anaconda Powershell Prompt
- Go to the unzipped directory
- Run the following commands:
conda create --name codiconda activate codiconda install pippip install -r requirements.txt
The CODI PPRL process depends on information pulled from a database structured to match the CODI Data Model. extract.py connects to a database and extracts information, cleaning and validating it to prepare it for the PPRL process. The script will output a temp-data/pii.csv file that contains the PII ready for garbling.
extract.py requires a database connection string to connect. Consult the SQLAlchemy documentation to determine the exact string for the database in use.
When finished, if you specify the --verbose flag, the script will print a report to the terminal, documenting various issues it found when extracting the data. An example execution of the script is included below:
$ python extract.py postgresql://codi:codi@localhost/codi -v
Total records exported: 5476
record_id
--------------------
given_name
--------------------
Contains Non-ASCII Characters: 226
Contains Non-printable Characters: 3
family_name
--------------------
Contains Non-ASCII Characters: 2
DOB
--------------------
sex
--------------------
phone_number
--------------------
household_street_address
--------------------
Contains Non-ASCII Characters: 1
Contains Non-printable Characters: 1
household_zip
--------------------
NULL Value: 9
parent_given_name
--------------------
Contains Non-ASCII Characters: 386
Contains Non-printable Characters: 4
parent_family_name
--------------------
NULL Value: 19
Contains Non-ASCII Characters: 4
parent_email
--------------------
NULL Value: 238
Contains Non-ASCII Characters: 12
clkhash will garble personally identifiable information (PII) in a way that it can be used for linkage later on. The CODI PPRL process garbles information a number of different ways. The garble.py script will manage executing clkhash multiple times and package the information for transmission to the DCC.
garble.py requires 3 different inputs:
- The location of a CSV file containing the PII to garble
- The location of a directory of clkhash linkage schema files
- The location of a secret file to use in the garbling process - this should be a text file containing a single hexadecimal string of at least 128 characters; the
testing-and-tuning/generate_secret.pyscript will create this for you if require it, e.g.:
python testing-and-tuning/generate_secret.py
This should create a new file called deidentification_secret.txt in your root directory.
garble.py requires that the location of the PII file, schema directory, and secret file are provided via positional arguments.
The clkhash schema files specify the fields that will be used in the hashing process as well as assigning weights to those fields. The example-schema directory contains a set of example schema that can be used to test the tools.
garble.py, and all other scripts in the repository, will provide usage information with the -h flag:
$ python garble.py -h
usage: garble.py [-h] [-o OUTPUTFILE] sourcefile schemadir secretfile
Tool for garbling PII in for PPRL purposes in the CODI project
positional arguments:
sourcefile Source PII CSV file
schemadir Directory of linkage schema
secretfile Location of de-identification secret file
optional arguments:
-h, --help show this help message and exit
-o OUTPUTFILE, --output OUTPUTFILE
Specify an output file. Default is output/garbled.zip
garble.py will package up the garbled PII files into a zip file called garbled.zip and place it in the output/ folder by default, you can change this with an --output flag if desired.
Example execution of garble.py is shown below:
$ python garble.py temp-data/pii.csv example-schema ../deidentification_secret.txt
CLK data written to output/name-sex-dob-phone.json
CLK data written to output/name-sex-dob-zip.json
CLK data written to output/name-sex-dob-parents.json
CLK data written to output/name-sex-dob-addr.json
Zip file created at: output/garbled.zip
You may now run households.py with the same arguments as the garble.py script, with the only difference being specifying a specific schema file instead of a schema directory - if no schema is specified it will default to the example-schema/household-schema/fn-phone-addr-zip.json (use -h flag for more information). NOTE: If you want to generate the testing and tuning files for development on a synthetic dataset, you need to specify the -t or --testrun flags
The households script will do the following:
- Attempt to group individuals into households and store those records in a csv file in temp-data
- Create a mapping file to be sent to the linkage agent, along with a zip file of household specific garbled information.
This information must be provided to the linkage agent if you would like to get a household linkages table as well.
Example run:
$ python households.py temp-data/pii.csv ../deidentification_secret.txt
Grouping individuals into households: 100%|███████████████████████| 819/819 [01:12<00:00, 11.37it/s]
CLK data written to output/households/fn-phone-addr-zip.json
Zip file created at: output/garbled_households.zip
Currently there is optional functionality for evaluation purposes to use blocking techniques to try and make the matching more efficient. After running garble.py you can run block.py to generate an additional blocking .zip file to send to the DCC / linkage agent.
Example run - note this is using the default settings, i.e. looking for the CLKs from the garble.py run in output/ and using the example-schema/blocking-schema/lambda.json LSH blocking configuration (Read more about blocking schmea here, and more about anonlink's LSH-based blocking approach here):
$ python block.py
Statistics for the generated blocks:
Number of Blocks: 79
Minimum Block Size: 1
Maximum Block Size: 285
Average Block Size: 31.10126582278481
Median Block Size: 9
Standard Deviation of Block Size: 59.10477331947379
Statistics for the generated blocks:
Number of Blocks: 82
Minimum Block Size: 1
Maximum Block Size: 232
Average Block Size: 29.963414634146343
Median Block Size: 9
Standard Deviation of Block Size: 45.5122952108199
Statistics for the generated blocks:
Number of Blocks: 75
Minimum Block Size: 1
Maximum Block Size: 339
Average Block Size: 32.76
Median Block Size: 10
Standard Deviation of Block Size: 61.43725430238738
Statistics for the generated blocks:
Number of Blocks: 80
Minimum Block Size: 1
Maximum Block Size: 307
Average Block Size: 30.7125
Median Block Size: 9
Standard Deviation of Block Size: 58.4333860157515
When anonlink matches across data owners / partners, it identifies records by their position in the file. It essentially uses the line number in the extracted PII file as the identifier for the record. When results are returned from the DCC, it will assign a LINK_ID to a line number in the PII CSV file.
To map the LINK_IDs back to PATIDs, use the linkid_to_patid.py script. The script takes two arguments:
- The path to the PII CSV file
- The path to the LINK_ID CSV file provided by the DCC
- [Optional] The path to the
temp-data/*_hid_mapping.csvfile created by thetesting-and-tuning/answer_key_map.pyscript (this should only be used if running with household linkage and testing against an answer key / ground truth) - [Optional] The path to the HOUSEHOLD_LINK_ID CSV file provided by the DCC if you provided household information
The script will create a file called linkid_to_patid.csv with the mapping of LINK_IDs to PATIDs in the output/ folder by default. If you are testing and running household linkage this will also create a linkid_to_hid.csv file in the output/ folder.
In between runs it is advisable to run rm temp-data/* to clean up temporary data files used for individuals runs.
The documentation above outlines the approach for a single data owner to run these tools. For a developer who is testing on a synthetic data set, they might want to run all of the above steps quickly and repeatedly for a list of artificial data owners.
In the linkage agent tools there is a Jupyter notebook under development that will run all of these steps through the notebook by invoking scripts in the testing-and-tuning/ folder.
If you would like to test household linkage you can currently run the garble.sh script (configuring the sites for which you have extracted pii). If you would like to test blocking you may run the blocking_garble.sh script. Note: for these scripts it is assumed that the pii files created by the extract.py have been renamed to their respective pii_{site}.csv.
Copyright 2020 The MITRE Corporation.
Approved for Public Release; Distribution Unlimited. Case Number 19-2008