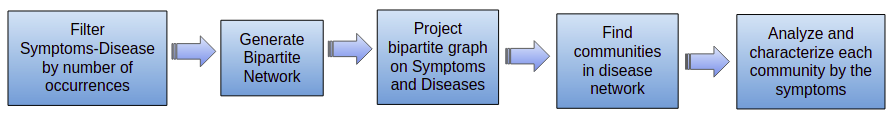
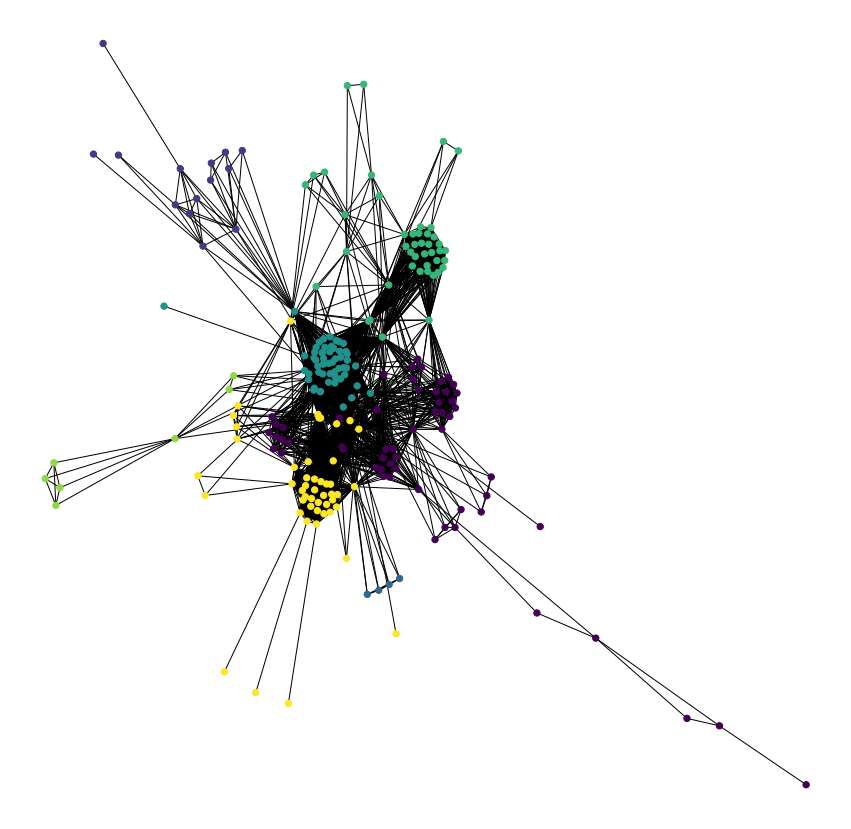
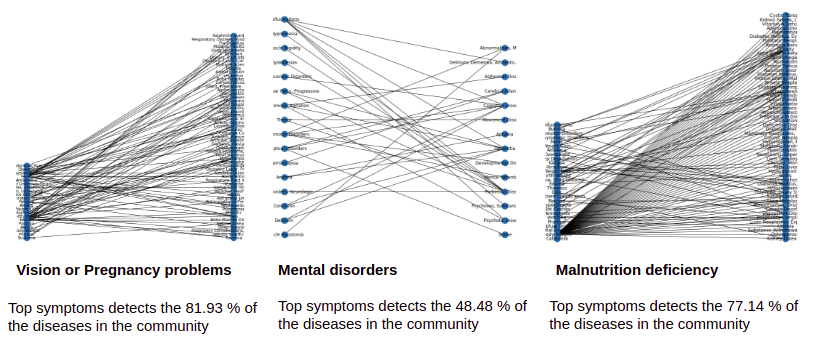
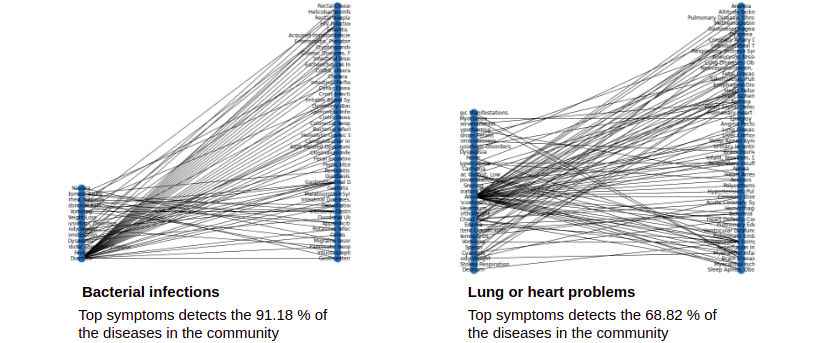
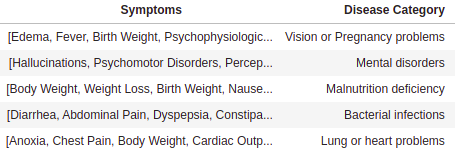
Analysis of the Symptoms-Disease Network database using communities.
The complete code of this project is in this repository, in the file: symptoms-disease-network.ipynb.
If the notebook does not load or takes a long time to load, please refresh the page.
The steps developed in this project were as follows:
The dataset was extracted from the paper: "Human symptoms–disease network" Link.
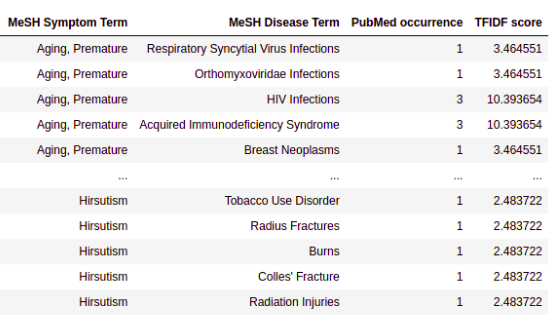
More exactly, here we employ the "Supplementary Data 3": Term co-occurrences between symptoms and diseases measured by TF-IDF weighted values. This table includes 147,978 records of symptom and disease relationships.
- Zhou, X., Menche, J., Barabási, AL. et al. Human symptoms–disease network. Nat Commun 5, 4212 (2014). https://doi.org/10.1038/ncomms5212
- Blondel, V. D., Guillaume, J.-L., Lambiotte, R., & Lefebvre, E. (2008). Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment, 2008(10), P10008. doi:10.1088/1742-5468/2008/10/p10008