All the code and datasets are avalable at this github repo
I am currently working on a workflow for Species Distribution Modeling using the targets R package. I’ve encountered two issues in a specific part of my workflow. Firstly, I am downloading presences in parallel using the crew package since the actual dataset consists of around 40,000 species. I have provided the relevant code below:
library(targets)
source("R/functions.R")
library(crew)
tar_option_set(packages = c("readr", "SDMWorkflows", "janitor", "data.table"),
controller = crew_controller_local(workers = 6),
error = "null")
list(
tar_target(file, "First_10_species.csv", format = "file"),
# Read the file
tar_target(data, get_data(file)),
# Filter the species to only plants
tar_target(Only_Plants, filter_plants(data)),
# Parallelize and retrieve species presences for species within Denmark
tar_target(Presences,
get_plant_presences(Only_Plants),
pattern = map(Only_Plants)),
# summarize the number of presences per species
tar_target(Presence_summary, summarise_presences(Presences),
pattern = map(Presences)),
# Filter to only the species that have 5 presences
tar_target(Over_5, Filter_Over_5(Presence_summary))
)The SDMWorkflows package is a package I made that you can install by using this code
remotes::install_github("Sustainscapes/SDMWorkflows")The accompanying function script (R/functions.R) is as follows:
get_data <- function(file) {
readr::read_csv(file) |>
janitor::clean_names()
}
filter_plants <- function(df){
result <- df |>
dplyr::filter(kingdom == "Plantae") |>
dplyr::pull(species) |>
unique() |>
head(10)
return(result)
}
get_plant_presences <- function(species){
SDMWorkflows::GetOccs(Species = unique(species),
WriteFile = FALSE,
Log = FALSE,
country = "DK",
limit = 100000,
year='1999,2023')
}
summarise_presences <- function(df){
Sum <- as.data.table(df)[, .N, keyby = .(family, genus, species)]
return(Sum)
}
Filter_Over_5 <- function(DT){
DT[N > 5]
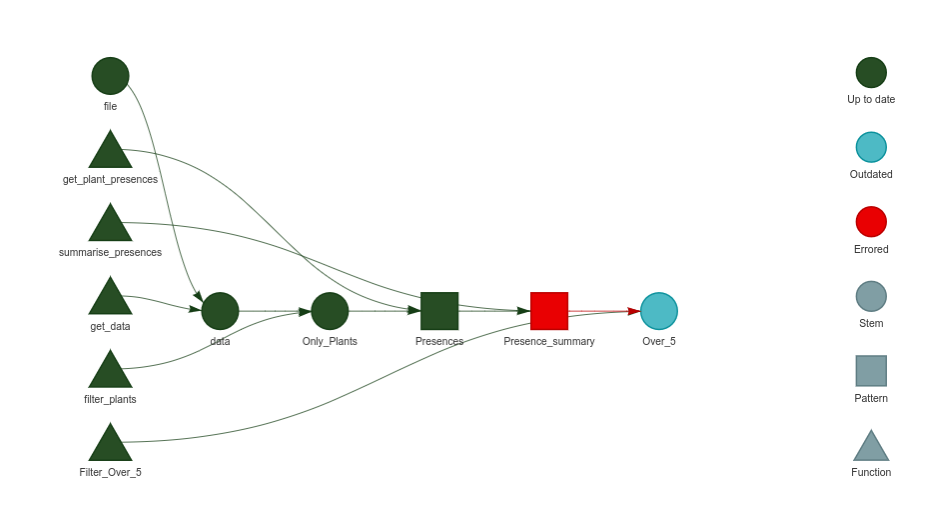
}While the workflow appears to be working well, some species summaries are showing errors. The errors are documented in the following table and figure
| name | error |
|---|---|
| Presence_summary_24c8afe2 | object genus not found |
| Presence_summary_7044ad96 | object genus not found |
| Presence_summary_a8f163ad | object genus not found |
| Presence_summary_c7ecffc9 | object genus not found |
knitr::include_graphics("PlotTarget.png")These errors are expected for species that did not present presences within Denmark. However, the summary appears fine, and from the initial 10 presences, it generates a data.table with 6 species, as illustrated in this table:
| family | genus | species | N |
|---|---|---|---|
| Pinaceae | Abies | Abies cephalonica | 1 |
| Pinaceae | Abies | Abies koreana | 3 |
| Pinaceae | Abies | Abies nordmanniana | 1130 |
| Pinaceae | Abies | Abies sibirica | 14 |
| Pinaceae | Abies | Abies veitchii | 2 |
| Thuidiaceae | Abietinella | Abietinella abietina | 9 |
I have two specific questions:
-
Addressing Errors in summarise_presences: Despite the errors, the results of summarise_presences are as expected. How can I eliminate these errors from the summary?
-
Filtering Species in Presences for Plotting: Suppose I want to use the results of Presences to plot coordinates with a function like PlotPres, but I only want to include species that appear in the Over_5 object. How can I achieve this mapping, considering that the species have names instead of branches?
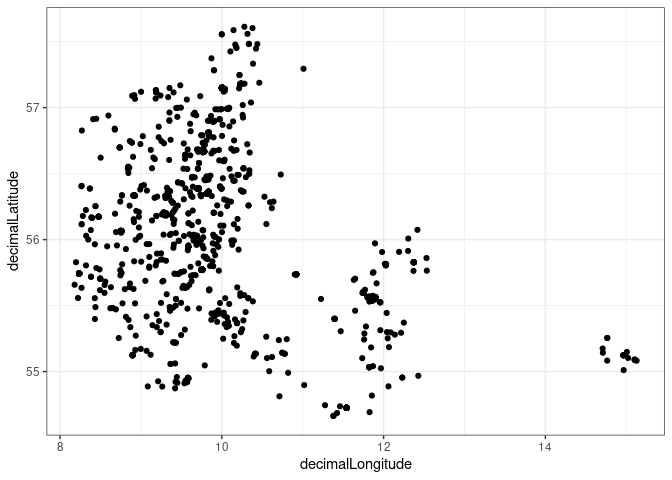
PlotPres <- function(df){
G <- ggplot(df , aes(x = decimalLongitude, y = decimalLatitude)) + geom_point() + theme_bw()
print(G)
}as you can see if I do this for branch 6 it works
PlotPres(tar_read("Presences", branches = 6)[[1]])