Method
This repository provides an implementation of the MGLasso (Multiscale
Graphical Lasso) algorithm: an approach for estimating sparse Gaussian
Graphical Models with the addition of a group-fused Lasso penalty.
MGLasso is described in the paper Inference of Multiscale Gaussian
Graphical Model.
MGLasso has these contributions:
-
We simultaneously infer a network and estimate a clustering structure by combining the neighborhood selection approach (Meinshausen and Bühlman, 2006) and convex clustering (Hocking et al. 2011).
-
We use a continuation with Nesterov smoothing in a shrinkage-thresholding algorithm (
CONESTA, Hadj-Selem et al. 2018) to solve the optimization problem.
To solve the MGLasso problem, we seek the regression vectors
that minimize
MGLasso package is based on the python implementation of the solver
CONESTA available in
pylearn-parsimony
library.
Package requirements and installation
- Install the
reticulatepackage and Miniconda if no conda distribution available on the OS.
install.packages('reticulate')
reticulate::install_miniconda()- Install
MGLasso, its python dependencies and configure the conda environmentrmglasso.
# install.packages('mglasso')
remotes::install_github("desanou/mglasso")library(mglasso)
install_pylearn_parsimony(envname = "rmglasso", method = "conda")
#> + "C:/Users/doedm/AppData/Local/r-miniconda/condabin/conda.bat" "install" "--yes" "--name" "rmglasso" "python=3.8"
reticulate::use_condaenv("rmglasso", required = TRUE)
reticulate::py_config()
#> python: C:/Users/doedm/AppData/Local/r-miniconda/envs/rmglasso/python.exe
#> libpython: C:/Users/doedm/AppData/Local/r-miniconda/envs/rmglasso/python38.dll
#> pythonhome: C:/Users/doedm/AppData/Local/r-miniconda/envs/rmglasso
#> version: 3.8.13 (default, Mar 28 2022, 06:59:08) [MSC v.1916 64 bit (AMD64)]
#> Architecture: 64bit
#> numpy: C:/Users/doedm/AppData/Local/r-miniconda/envs/rmglasso/Lib/site-packages/numpy
#> numpy_version: 1.22.4
#> conesta_solver: [NOT FOUND]
#>
#> NOTE: Python version was forced by use_python functionThe conesta_solver is delay loaded. See reticulate::import_from_path
for details.
An example of use is given below.
Illustration on a simple model
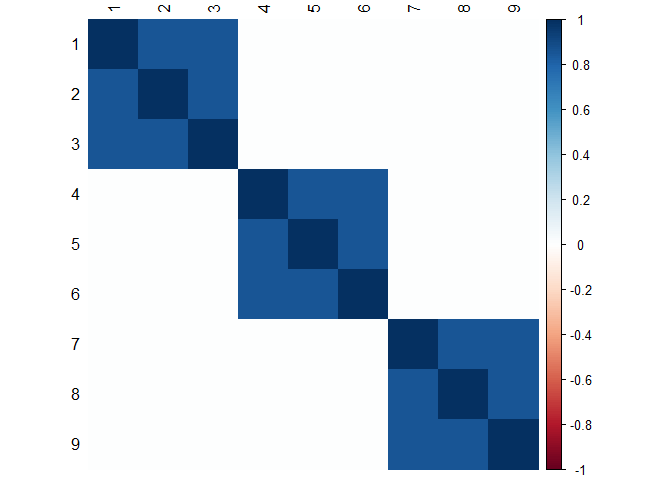
Simulate a block diagonal model
We simulate a
-block
diagonal model where each block contains
variables. The intra-block correlation level is set to
while the correlations outside the blocks are kept to
.
library(Matrix)
n = 50
K = 3
p = 9
rho = 0.85
blocs <- list()
for (j in 1:K) {
bloc <- matrix(rho, nrow = p/K, ncol = p/K)
for(i in 1:(p/K)) { bloc[i,i] <- 1 }
blocs[[j]] <- bloc
}
mat.correlation <- Matrix::bdiag(blocs)
corrplot::corrplot(as.matrix(mat.correlation), method = "color", tl.col="black")Simulate gaussian data from the covariance matrix
set.seed(11)
X <- mvtnorm::rmvnorm(n, mean = rep(0,p), sigma = as.matrix(mat.correlation))
colnames(X) <- LETTERS[1:9]Run mglasso()
We set the sparsity level
to
and rescaled it with the size of the sample.
X <- scale(X)
res <- mglasso(X, lambda1 = 0.2*n, lambda2_start = 0.1, fuse_thresh = 1e-3, verbose = FALSE)To launch a unique run of the objective function call the conesta
function.
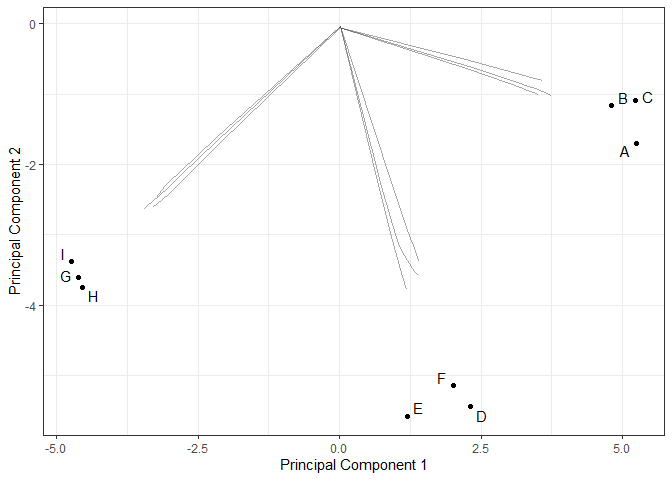
temp <- mglasso::conesta(X, lam1 = 0.2*n, lam2 = 0.1)Estimated clustering path
We plot the clustering path of mglasso method on the 2 principal
components axis of
.
The path is drawn on the predicted
’s.
library(ggplot2)
library(ggrepel)
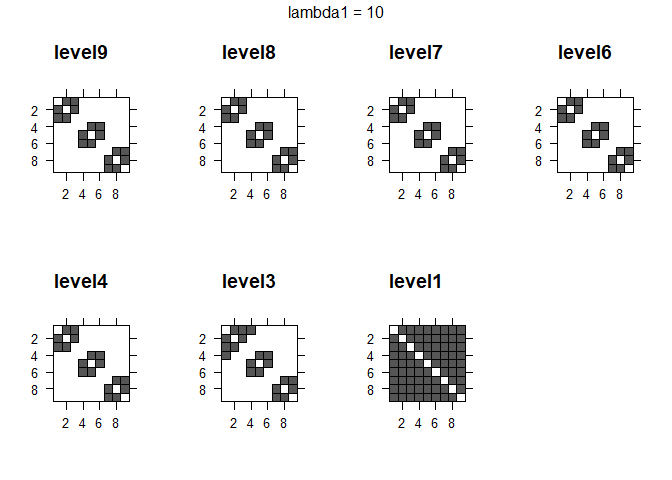
mglasso:::plot_clusterpath(as.matrix(X), res)Estimated adjacency matrices along the clustering path
As the the fusion penalty increases from level9 to level1 we observe
a progressive fusion of adjacent edges.
plot_mglasso(res)Reference
Edmond, Sanou; Christophe, Ambroise; Geneviève, Robin; (2022): Inference of Multiscale Gaussian Graphical Model. ArXiv. Preprint. https://doi.org/10.48550/arXiv.2202.05775