Fast smoothing of per-vertex data on triangular meshes for Python.
This package package performs smoothing of per-vertex data on triangular meshes. Such smoothing is typically used to reduce high-frequency noise and improve signal-to-noise ration (SNR). The algorithm for iterative nearest-neighbor smoothing is trivial, but involves nested tight loops, which are very slow in Python, so this package calls into C++ via pybind11 to achieve high performance.
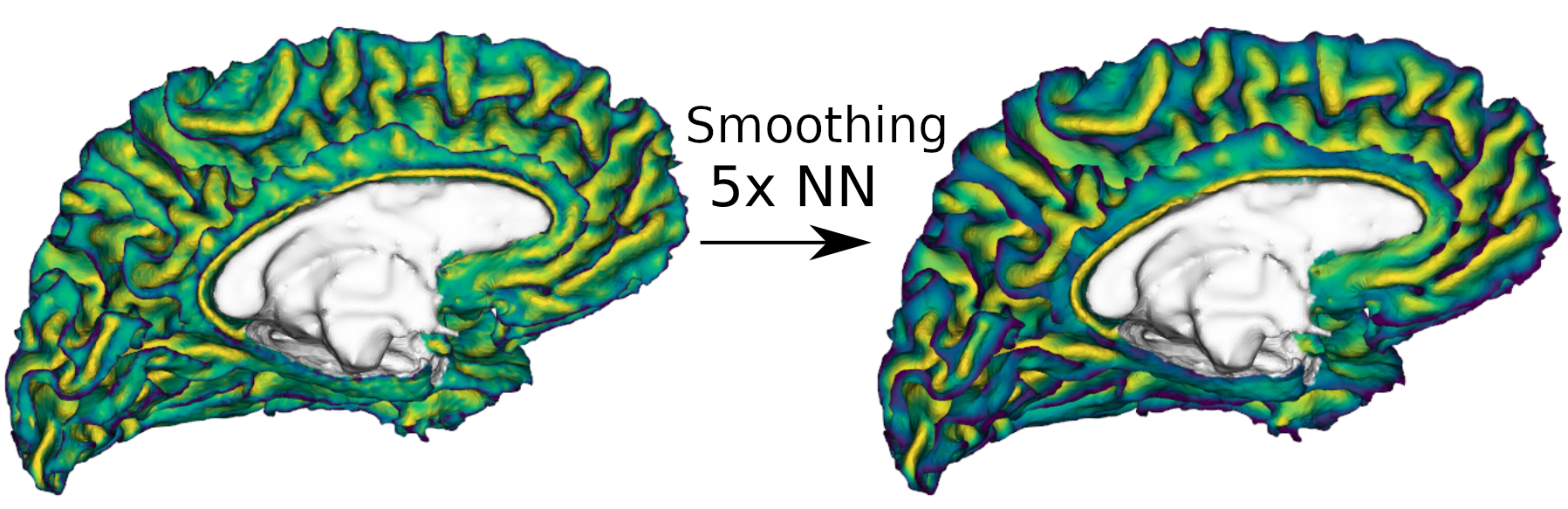
Fig.1: Noisy per-vertex data on a brain mesh (left), and the same data after smoothing (right). White represents NA values. The data, mapped to the colors of the viridis colormap in the visualization above, represents the vertex-wise mean curvature of the mesh in this example, but it could be anything one can map to a mesh, one value per vertex.
This is a Python implementation of the haze package for R. The haze website offers a more detailed explanation of the motivation and use cases.
Via pip:
pip install pyhazeAlternatively, if you want to use conda:
conda install -c dfspspirit pyhazeHere is a simple example using the pyhaze.smooth_pvd function.
import pyhaze
import numpy as np
vertices, faces = pyhaze.construct_cube()
pvd_data = np.arange(len(vertices), dtype=float)
smoothed_data = pyhaze.smooth_pvd(vertices, faces, pvd_data.tolist(), num_iter=5)A note on the mesh representation used, so you can replace the vertices and faces with your own triangular mesh:
verticesis a list of lists offloat, with dimensionN, 3forNvertices. So the outer list has lengthN. The 3 columns (length of all inner lists) are the x,y,z coordinates for each vertex.facesis a list of lists ofint, with dimensionM, 3forMfaces. So the outer list has lengthM. The 3 columns (length of all inner lists) are the 3 vertices (given as indices intovertices) making up the respective triangular face.
For very large meshes, it pays off to pre-compute the adjacency list of the mesh with a fast method, such as with the igl Python package, which provides Python bindings for libigl, and use the pyhaze.smooth_pvd_adj function.
import pyhaze
import numpy as np
import igl
vertices, faces = pyhaze.construct_cube()
pvd_data = np.arange(len(vertices), dtype=float)
faces_igl = np.array(faces).reshape(-1, 3).astype(np.int64)
mesh_adj = igl.adjacency_list(faces_igl) # Compute adjacency list with igl.
res = pyhaze.smooth_pvd_adj(mesh_adj, pvd_data.tolist(), num_iter=5)See the unit tests for more examples.
pyhaze was written by Tim Schäfer