This open-source project contains the Pytorch implementation of our approach (ConEx), training and evaluation scripts. We added ConEx and its variant AConEx into DICE Embeddings Framework open-source project to ease the deployment and the distributed computing. Therein, we provided pre-trained models on many knowledge graphs
In the below, we provide a brief overview of the link prediction results. Results are sorted in descending order of the size of the respective dataset.
| MRR | Hits@10 | Hits@3 | Hits@1 | |
|---|---|---|---|---|
| DistMult | .340 | .540 | .380 | .240 |
| ComplEx | .360 | .550 | .400 | .260 |
| ConvE | .400 | .620 | .490 | .350 |
| HypER | .530 | .680 | .580 | .460 |
| RotatE | .500 | .670 | .550 | .400 |
| ConEx | .553 | .696 | .601 | .477 |
(*) denotes the newly reported link prediction results.
| MRR | Hits@10 | Hits@3 | Hits@1 | |
|---|---|---|---|---|
| DistMult | .241 | .419 | .263 | .155 |
| ComplEx | .247 | .428 | .275 | .158 |
| ConvE | .335 | .501 | .356 | .237 |
| RESCAL* | .357 | .541 | .393 | .263 |
| DistMult* | .343 | .531 | .378 | .250 |
| ComplEx* | .348 | .536 | .384 | .253 |
| ConvE* | .339 | .521 | .369 | .248 |
| HypER | .341 | .520 | .376 | .252 |
| NKGE | .330 | .510 | .365 | .241 |
| RotatE | .338 | .533 | .375 | .241 |
| TuckER | .358 | .544 | .394 | .266 |
| QuatE | .366 | .556 | .401 | .271 |
| ConEx | .366 | .555 | .403 | .271 |
| Ensemble.ConEx | .376 | .570 | .415 | .279 |
(*) denotes the newly reported link prediction results.
| MRR | Hits@10 | Hits@3 | Hits@1 | |
|---|---|---|---|---|
| DistMult | .430 | .490 | .440 | .390 |
| ComplEx | .440 | .510 | .460 | .410 |
| ConvE | .430 | .520 | .440 | .400 |
| RESCAL* | .467 | .517 | .480 | .439 |
| DistMult* | .452 | .530 | .466 | .413 |
| ComplEx* | .475 | .547 | .490 | .438 |
| ConvE* | .442 | .504 | .451 | .411 |
| HypER | .465 | .522 | .477 | .436 |
| NKGE | .450 | .526 | .465 | .421 |
| RotatE | .476 | .571 | .492 | .428 |
| TuckER | .470 | .526 | .482 | .443 |
| QuatE | .482 | .572 | .499 | .436 |
| ConEx | .481 | .550 | .493 | .448 |
| Ensemble.ConEx | .485 | .559 | .495 | .450 |
We spot flaws on WN18RR, FB15K-237 and YAGO3-10. More specifically, the validation and test splits of the dataset contain entities that do not occur in the training split. We refer Out-of-Vocabulary Entities in Link Prediction for more details.
| MRR | Hits@10 | Hits@3 | Hits@1 | |
|---|---|---|---|---|
| DistMult-ComplEx | .475 | .579 | .497 | .426 |
| DistMult-TuckER | .476 | .569 | .492 | .433 |
| ConEx-DistMult | .484 | .580 | .501 | .439 |
| ConEx-ComplEx | .501 | .589 | .518 | .456 |
| ConEx-TuckER | .514 | .583 | .526 | .479 |
| Ensemble.ConEx | .517 | .594 | .526 | .479 |
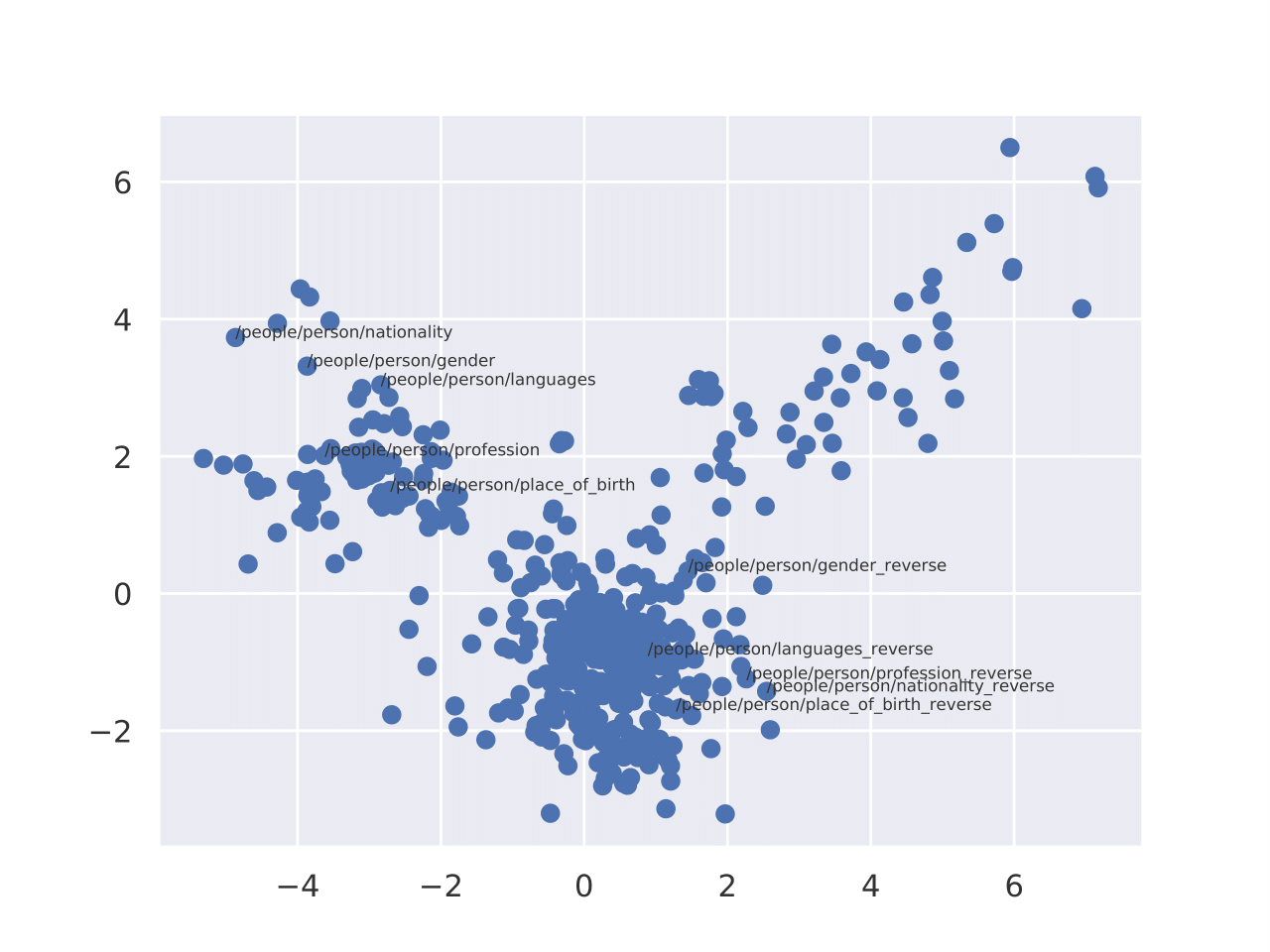
A 2D PCA projection of relation embeddings on the FB15K-237 dataset. The Figure shows that inverse relations cluster in distant regions. Note that we applied the standard data augmentation technique
To generate inverse relations, relations are renamed by adding suffix of inverse as done in~\cite{balavzevic2019tucker}.
First clone the repository:
git clone https://github.com/dice-group/Convolutional-Complex-Knowledge-Graph-Embeddings.git
Then obtain the required libraries:
conda env create -f environment.yml
source activate conex
The code is compatible with Python 3.6.4.
run_script.pycan be used to train ConEx on a desired dataset.grid_search.pycan be used to rerun our experiments.
Please contact: caglar.demir@upb.de, if you wish to obtain ConEx embeddings of specific dataset.
- Forte embeddings
- Hepatitis embeddings
- Lymphography embeddings
- Mammographic embeddings
- Animals embeddings
- YAGO3-10 embeddings
- FB15K-237 embeddings
- FB15K embeddings
- WN18RR embeddings
- WN18 embeddings
Please follow the next steps to reproduce all reported results.
- Unzip the datasets:
unzip KGs.zip - Create a folder for pretrained models:
mkdir PretrainedModels - Download pretrained models via hobbitdata into
PretrainedModels. python reproduce_lp.pyreproduces link prediction results on the FB15K-237, FB15K, WN18, WN18RR and YAGO3-10 benchmark datasets.python reproduce_baselines.pyreproduces link prediction results of DistMult, ComplEx and TuckER on the FB15K-237, WN18RR and YAGO3-10 benchmark datasets.settings.jsonfiles store the hyperparameter setting for each model.python reproduce_ensemble.pyreports link prediction results of ensembled models.python reproduce_lp_new.pyreports link prediction results on WN18RR*, FB15K-237* and YAGO3-10*.python reproduce_ablation.py.pyreports link prediction results of our ablation study.
In the below, we provide a brief overview of the link prediction results.
| MRR | Hits@10 | Hits@3 | Hits@1 | |
|---|---|---|---|---|
| DistMult | .340 | .540 | .380 | .240 |
| ComplEx | .360 | .550 | .400 | .260 |
| ConvE | .440 | .620 | .490 | .350 |
| HypER | .530 | .678 | .580 | .455 |
| RotatE | .495 | .670 | .550 | .400 |
| DistMult | .543 | .683 | .590 | .466 |
| ComplEx | .547 | .690 | .594 | .468 |
| TuckER | .427 | .609 | .476 | .331 |
| ConEx | .553 | .696 | .601 | .474 |
| MRR | Hits@10 | Hits@3 | Hits@1 | |
|---|---|---|---|---|
| DistMult | .241 | .419 | .263 | .155 |
| ComplEx | .247 | .428 | .275 | .158 |
| ConvE | .335 | .501 | .356 | .237 |
| DistMult | .343 | .531 | .378 | .250 |
| ComplEx | .348 | .536 | .384 | .253 |
| ConvE | .339 | .521 | .369 | .248 |
| RotatE | .338 | .533 | .375 | .241 |
| HypER | .341 | .520 | .376 | .252 |
| DistMult | .353 | .539 | .390 | .260 |
| ComplEx | .332 | .509 | .366 | .244 |
| TuckER | .363 | .553 | .400 | .268 |
| ConEx | .366 | .555 | .403 | .271 |
| Ensemble of ConEx | .376 | .570 | .415 | .279 |
We based our implementation on the open source implementation of TuckER. We would like to thank for the readable codebase.
@inproceedings{demir2021convolutional,
title={Convolutional Complex Knowledge Graph Embeddings},
author={Caglar Demir and Axel-Cyrille Ngonga Ngomo},
booktitle={Eighteenth Extended Semantic Web Conference - Research Track},
year={2021},
url={https://openreview.net/forum?id=6T45-4TFqaX}}
For any further questions or suggestions, please contact: caglar.demir@upb.de or caglardemir8@gmail.com