InfIntE stands for Inference of Interactions using Explainable machine learning. This package uses abundance data to directly infer ecological interactions using PyGol, an Abductive/Inductive logic program, classified by their interaction type. For a detailed explanation of the InfIntE procedure and its implementation, please see:
Barroso-Bergada D, Tamaddoni-Nezhad A, Varghese D, Vacher C, Galic N, Laval V, Suffert F, Bohan DA (2023). “Unravelling the web of dark interactions: Explainable inference of the diversity of microbial interactions.” In Advances in Ecological Research: Roadmaps: Part A, volume 68, 155-183. Academic Press. https://doi.org/10.1016/bs.aecr.2023.09.005.
InfIntE and required packages are installed using devtools
library(devtools)
if(!"InfIntE" %in% rownames(installed.packages())){
install_github("didacb/InfIntE")
}Interaction inference uses the logical inference process of abduction. Abduction is performed using PyGol. PyGol is written in c. To compile PyGol and obtain the functions for abduction run:
load_PyGol()To use PyGol it is necessary that the following python modules are installed (preferably using pip3):
- numpy
- texttable
- cython
Windows users also need to install the Microsoft C++ Build Tools
We illustrate how InfIntE works using wheat foliar bacterial ASV data. The data characteristics are detailed here. The ASV data is in phyloseq format. First, let’s import and subset the data to obtain a manageable size.
#Import data
library(InfIntE)
library(phyloseq)
data("BCM_16S_wheat_phyloseq_filtered_lulu")
wheat_metadata<- sample_data(BCM_16S_wheat_phyloseq_filtered_lulu)
#Keep only green samples from march
selected_samples<- wheat_metadata$Date == "03_18" &
wheat_metadata$Specie == "wheat" &
wheat_metadata$Variety == "Apa" &
wheat_metadata$Tissue == "G"
asv_subset<- prune_samples(selected_samples, BCM_16S_wheat_phyloseq_filtered_lulu)
#Obtain the sequencing depth for the whole dataset
depth<- sample_sums(asv_subset)
#Keep only the most abundant ASVs
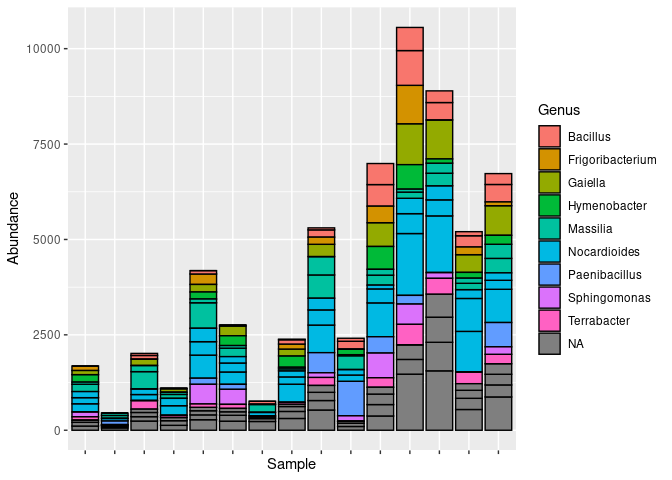
asv_subset<- prune_taxa(taxa_sums(asv_subset)>2000, asv_subset)The wheat bacterial community has many different bacterial genus represented
library(ggplot2)
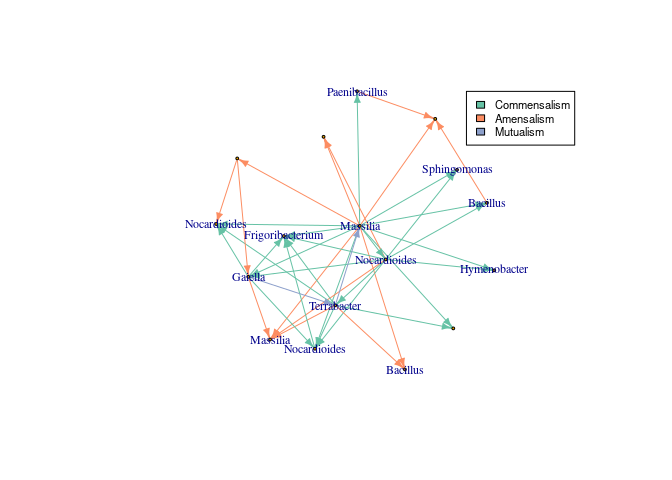
plot_bar(asv_subset, fill = "Genus")+theme(axis.text.x = element_blank())To infer interactions, InfIntE offers a homonymous function to perform the whole pipeline, from an OTU abundance table to abduced ecological interactions, in a single run.
library(igraph)
#Infer interactions using the complete sequencing depth
interactions<- infinte(otu_tb = data.frame(otu_table(asv_subset, taxa_are_rows = T)),
exclusion = TRUE, ncores = 1, nperms = 50, depth = depth)
#Get network
network_graph<-graph_from_data_frame(interactions$selected_interactions)
#Change ASV names to genus
V(network_graph)$name<- data.frame(tax_table(asv_subset))[V(network_graph)$name,]$Genus
#Add color to different interactions
library(RColorBrewer)
colors_edges<- brewer.pal(5, "Set2")
E(network_graph)$color<- sapply(E(network_graph)$lnk, function(x){
colors_edges[which(unique(E(network_graph)$lnk)==x)]
})
#Plot
lay <- layout.kamada.kawai(network_graph)
set.seed(123)
plot(network_graph, layout=lay, vertex.size=2,
vertex.label.cex = 0.75, edge.arrow.size=0.5)
#Add legend
legend(0.8,1, legend=unique(E(network_graph)$lnk),
fill=unique(E(network_graph)$color), cex=0.7)But, what does InfIntE do, exactly, to learn interactions? InfIntE uses an hypothesis of interaction written as a logical relation between OTU/ASV presence, abundance and effects, as changes in OUT/ASV abundance.
hypothesis<-
c("abundance(C1,C2,S1,up):-presence(C2,S2,yes)&presence1(C1,S2,no)&effect_up(S2,S1)",
"abundance(C1,C2,S1,app):-presence(C2,S2,yes)&presence1(C1,S2,no)&effect_up(S2,S1)",
"abundance(C1,C2,S1,down):-presence(C2,S2,yes)&presence1(C1,S2,no)&effect_down(S2,S1)",
"abundance(C1,C2,S1,dis):-presence(C2,S2,yes)&presence1(C1,S2,no)&effect_down(S2,S1)")It then transforms the ASV matrix into logic clauses related by the hypothesis
# Join absolute and compositional data in a table
otu_data <- join_abundances(otu_tb=otu_table(asv_subset, taxa_are_rows = T),
absolute_abundance = NULL, depth = depth)
# All possible pairs of samples
comparisons <- get_comparsions(length(otu_data$samp_names))
# Get head logic clauses
head_clauses <- lapply(rownames(otu_data$otu_tb), function(otu) {
pos <- which(rownames(otu_data$otu_tb) == otu)
abundances <- do.call(
what = otu_data$abundance_function[pos],
args = list(
"otu_abundance" = otu_data$otu_tb[pos, , drop = FALSE],
"comparisons" = comparisons, "depth" = otu_data$depth, "exclusion" = TRUE
)
)
return(abundances)
})
head_clauses <- unlist(head_clauses)
# Get Body logic clauses
body_clauses <- get_presence(otu_data)
head(body_clauses)## [1] "presence(c1,s1,yes)." "presence(c2,s1,no)." "presence(c3,s1,no)."
## [4] "presence(c4,s1,no)." "presence(c5,s1,yes)." "presence(c6,s1,no)."
Then, PyGol is used to generate the bottom clause and abduce the effects on the OTU abundance caused by other ASVs. InfIntE renames the ASVs during the abduction to optimize process.
# Produce bottom clause
bottom_clauses <- get_bottom_clause(otu_data = otu_data,
head_clauses = head_clauses,
body_clauses = body_clauses)
# Abduce effects
abduced_effects <- abduce(bottom = bottom_clauses, hypothesis = hypothesis)
# Get I values
abduced_effects <- get_I_values(abduced_effects)#Infer interactions
head(abduced_effects)## sp1 sp2 lnk comp
## 1 s1 s1 effect_up 2542
## 2 s1 s10 effect_up 7
## 3 s1 s11 effect_down 0
## 4 s1 s12 effect_down 540
## 5 s1 s13 effect_up 363
## 6 s1 s14 effect_down 45
To select interactions, InfIntE uses the pulsar package to run the StARS model selection.
# Length observations
mx <- length(bottom_clauses$head)
# Lambda distribution
lambda <- pulsar::getLamPath(max = mx, min = 0, 50, FALSE)
# Pulsar execution
pulsar_output <- pulsar::pulsar(t(otu_data$otu_tb),
fun = pulsar_infinte,
fargs = list(lambda = lambda, bottom_clauses = bottom_clauses,
hypothesis = hypothesis, exclusion = TRUE, otu_data = otu_data),
rep.num = 50, lb.stars = TRUE, ub.stars = TRUE, thresh = 0.01, ncores = 25,
)
# Format output to dataframe
fitted_model <- pulsar::refit(pulsar_output, criterion = "stars")
interactions <- data.frame(igraph::get.edgelist(
igraph::graph_from_adjacency_matrix(fitted_model$refit$stars)))
head(interactions)## X1 X2
## 1 s10 s1
## 2 s11 s1
## 3 s16 s1
## 4 s4 s1
## 5 s8 s1
## 6 s16 s10
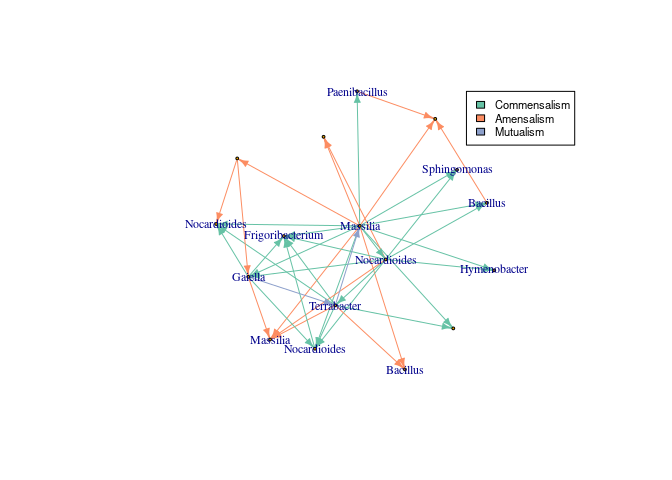
As a final step, InfIntE classifies the interactions by their type.
# Take values from abduced effects dataframe
interactions <- abduced_effects[paste0(abduced_effects[, 1], abduced_effects[, 2])
%in% paste0(interactions[, 1], interactions[, 2]), ]
# Classify and give back original names
interactions <- classify_interactions(interactions)
interactions <- return_names(interactions, otu_data$otu_names)
#Get network
network_graph<-graph_from_data_frame(interactions)
#Change ASV names to genus
V(network_graph)$name<- data.frame(tax_table(asv_subset))[V(network_graph)$name,]$Genus
#Add color to different interactions
colors_edges<- brewer.pal(5, "Set2")
E(network_graph)$color<- sapply(E(network_graph)$lnk, function(x){
colors_edges[which(unique(E(network_graph)$lnk)==x)]
})
#Plot
lay <- layout.kamada.kawai(network_graph)
set.seed(123)
plot(network_graph, layout=lay, vertex.size=2,
vertex.label.cex = 0.75, edge.arrow.size=0.5 )
#Add legend
legend(0.8,1, legend=unique(E(network_graph)$lnk),
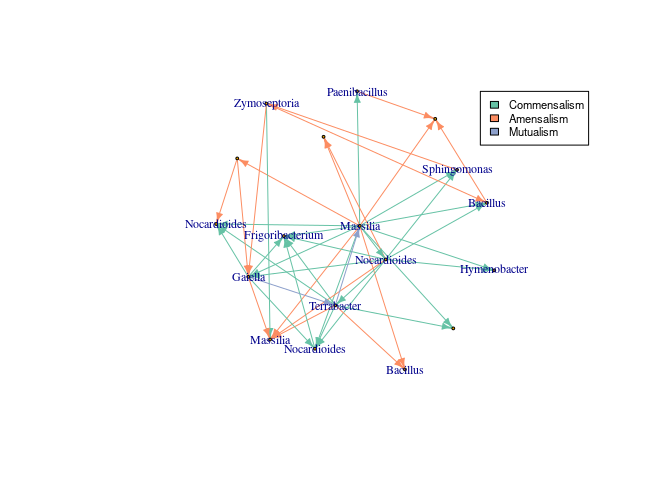
fill=unique(E(network_graph)$color), cex=0.7)InfIntE can also use absolute abundance data, complementing the compositional data obtained from eDNA. In this example we use the qPCR measurements of the pathogen Z. tritici available in the metadata.
#Retrieve absolute abundance
absolute_abundance<- t(data.frame(sample_data(asv_subset))[,7,drop=FALSE])
absolute_abundance<- ifelse(is.na(absolute_abundance),0,absolute_abundance)
#Infer interactions
interactions_z<- infinte(otu_tb = otu_table(asv_subset,
taxa_are_rows = T), ncores = 25,
absolute_abundance = absolute_abundance, exclusion = TRUE, depth = depth)
network_graph_z<-graph_from_data_frame(interactions_z$selected_interactions)
#Change Zymoseptoria ASV names to genus
zymo.pos<- grep("Zymoseptoria", V(network_graph_z)$name)
V(network_graph_z)$name<- data.frame(tax_table(asv_subset))[V(network_graph_z)$name,]$Genus
V(network_graph_z)$name[zymo.pos]<- "Zymoseptoria"
#Add color to different interactions
colors_edges<- brewer.pal(5, "Set2")
E(network_graph_z)$color<- sapply(E(network_graph_z)$lnk, function(x){
colors_edges[which(unique(E(network_graph_z)$lnk)==x)]})
#Reorder the layout to obtain the same vertex position
lay.zym <- rbind(lay, c(-1.5,3.5))
lay.zym<- lay.zym[c(1,2,3,4,5,18,13,6,7,8,9,10,11,12,14,15,16,17),]
set.seed(123)
plot(network_graph_z, layout=lay.zym, vertex.size=2,
vertex.label.cex = 0.75, edge.arrow.size=0.5 )
#Add legend
legend(0.9,1, legend=unique(E(network_graph_z)$lnk),
fill=unique(E(network_graph_z)$color), cex=0.7)