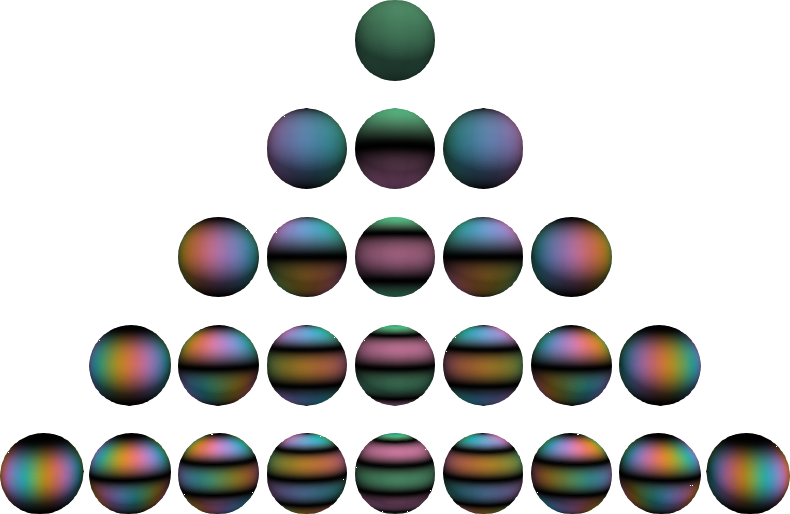All about orthogonal polynomials.
orthopy provides various orthogonal polynomial classes for lines, triangles, disks, spheres, n-cubes, the nD space with weight function exp(-r2) and more. All computations are done using numerically stable recurrence schemes. Furthermore, all functions are fully vectorized and can return results in exact arithmetic.
Install orthopy from PyPi via
pip install orthopy
The main function of all submodules is the iterator Eval which evaluates the series of
orthogonal polynomials with increasing degree at given points using a recurrence
relation, e.g.,
import orthopy
x = 0.5
evaluator = orthopy.c1.legendre.Eval(x, "classical")
for _ in range(5):
print(next(evaluator))1.0 # P_0(0.5)
0.5 # P_1(0.5)
-0.125 # P_2(0.5)
-0.4375 # P_3(0.5)
-0.2890625 # P_4(0.5)Other ways of getting the first n items are
evaluator = Eval(x, "normal")
vals = [next(evaluator) for _ in range(n)]
import itertools
vals = list(itertools.islice(Eval(x, "normal"), n))Instead of evaluating at only one point, you can provide any array for x; the
polynomials will then be evaluated for all points at once. You can also use sympy for
symbolic computation:
import itertools
import orthopy
import sympy
x = sympy.Symbol("x")
evaluator = orthopy.c1.legendre.Eval(x, "classical")
for val in itertools.islice(evaluator, 5):
print(sympy.expand(val))1
x
3*x**2/2 - 1/2
5*x**3/2 - 3*x/2
35*x**4/8 - 15*x**2/4 + 3/8
All Eval methods have a scaling argument which can have three values:
"monic": The leading coefficient is 1."classical": The maximum value is 1 (or (n+alpha over n))."normal": The integral of the squared function over the domain is 1.
For univariate ("one-dimensional") integrals, every new iteration contains one function. For bivariate ("two-dimensional") domains, every level will contain one function more than the previous, and similarly for multivariate families. See the tree plots below.
| Legendre | Chebyshev 1 | Chebyshev 2 |
Jacobi, Gegenbauer (α=β), Chebyshev 1 (α=β=-1/2), Chebyshev 2 (α=β=1/2), Legendre (α=β=0) polynomials.
import orthopy
orthopy.c1.legendre.Eval(x, "normal")
orthopy.c1.chebyshev1.Eval(x, "normal")
orthopy.c1.chebyshev2.Eval(x, "normal")
orthopy.c1.gegenbauer.Eval(x, "normal", lmbda)
orthopy.c1.jacobi.Eval(x, "normal", alpha, beta)The plots above are generated with
import orthopy
orthopy.c1.jacobi.show(5, "normal", 0.0, 0.0)
# plot, savefig also existRecurrence coefficients can be explicitly retrieved by
import orthopy
rc = orthopy.c1.jacobi.RecurrenceCoefficients(
"monic", # or "classical", "normal"
alpha=0, beta=0, symbolic=True
)
print(rc.p0)
for k in range(5):
print(rc[k])1
(1, 0, None)
(1, 0, 1/3)
(1, 0, 4/15)
(1, 0, 9/35)
(1, 0, 16/63)
(Generalized) Laguerre polynomials.
evaluator = orthopy.e1r.Eval(x, alpha=0, scaling="normal")Hermite polynomials come in two standardizations:
"physicists"(against the weight functionexp(-x ** 2)"probabilists"(against the weight function1 / sqrt(2 * pi) * exp(-x ** 2 / 2)
evaluator = orthopy.e1r2.Eval(
x,
"probabilists", # or "physicists"
"normal"
)Not all of those are polynomials, so they should really be called associated Legendre functions. The kth iteration contains 2k+1 functions, indexed from -k to k. (See the color grouping in the above plot.)
evaluator = orthopy.c1.associated_legendre.Eval(
x, phi=None, standardization="natural", with_condon_shortley_phase=True
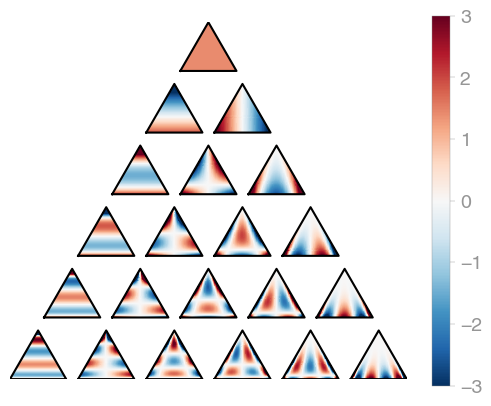
)orthopy's triangle orthogonal polynomials are evaluated in terms of barycentric
coordinates, so the
X.shape[0] has to be 3.
import orthopy
bary = [0.1, 0.7, 0.2]
evaluator = orthopy.t2.Eval(bary, "normal")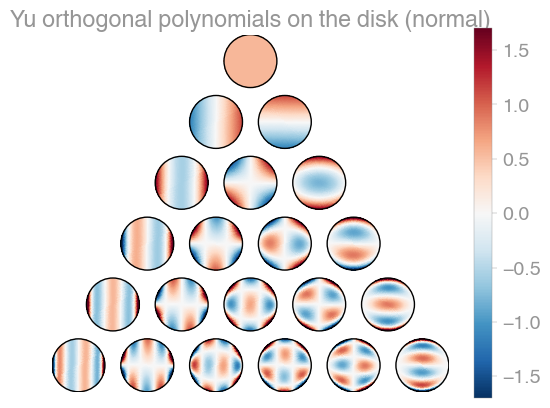 |
 |
 |
|---|---|---|
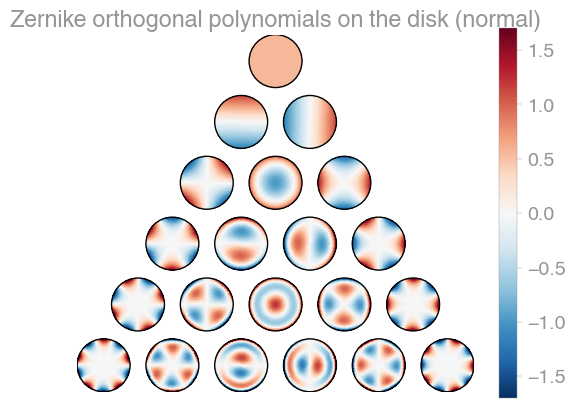
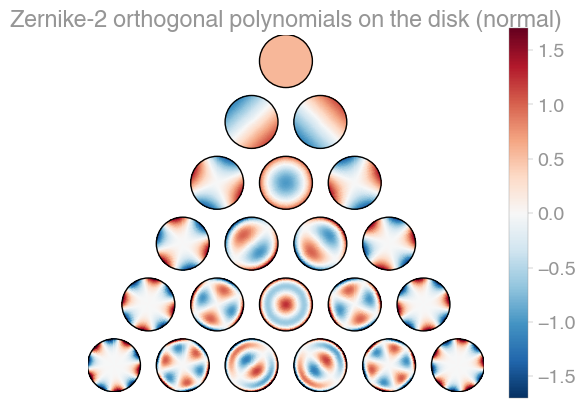
| Xu | Zernike | Zernike 2 |
orthopy contains several families of orthogonal polynomials on the unit disk: After Xu, Zernike, and a simplified version of Zernike polynomials.
import orthopy
x = [0.1, -0.3]
evaluator = orthopy.s2.xu.Eval(x, "normal")
# evaluator = orthopy.s2.zernike.Eval(x, "normal")
# evaluator = orthopy.s2.zernike2.Eval(x, "normal")Complex-valued spherical harmonics, plotted with cplot coloring (black=zero, green=real positive, pink=real negative, blue=imaginary positive, yellow=imaginary negative). The functions in the middle are real-valued. The complex angle takes n turns on the nth level.
evaluator = orthopy.u3.EvalCartesian(
x,
scaling="quantum mechanic" # or "acoustic", "geodetic", "schmidt"
)
evaluator = orthopy.u3.EvalSpherical(
theta_phi, # polar, azimuthal angles
scaling="quantum mechanic" # or "acoustic", "geodetic", "schmidt"
)To generate the above plot, write the tree mesh to a file
import orthopy
orthopy.u3.write_tree("u3.vtk", 5, "quantum mechanic")and open it with ParaView. Select the srgb1 data set and turn off Map Scalars.
 |
 |
|
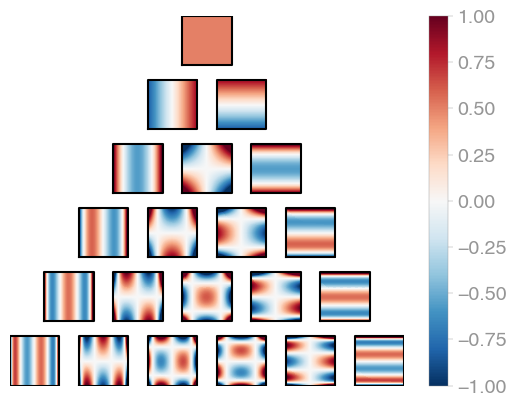
|---|---|---|
| C1 (Legendre) | C2 | C3 |
Jacobi product polynomials.
All polynomials are normalized on the n-dimensional cube. The dimensionality is
determined by X.shape[0].
evaluator = orthopy.cn.Eval(X, alpha=0, beta=0)
values, degrees = next(evaluator)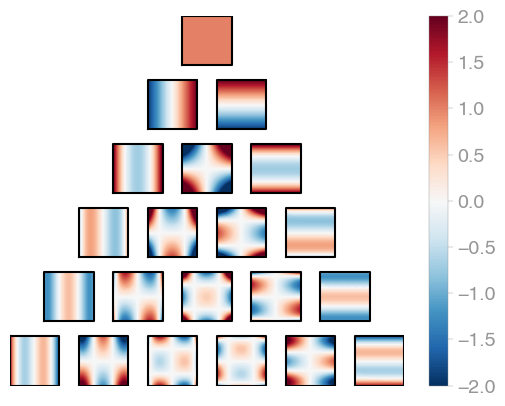 |
 |
|
|---|---|---|
| E1r2 | E2r2 | E3r2 |
Hermite product polynomials.
All polynomials are normalized over the measure. The dimensionality is determined by
X.shape[0].
evaluator = orthopy.enr2.Eval(
x,
standardization="probabilists" # or "physicists"
)
values, degrees = next(evaluator)-
Generating recurrence coefficients for 1D domains with Stieltjes, Golub-Welsch, Chebyshev, and modified Chebyshev.
-
The the sanity of recurrence coefficients with test 3 from Gautschi's article: computing the weighted sum of orthogonal polynomials:
orthopy.tools.gautschi_test_3(moments, alpha, beta)
-
Clenshaw algorithm for computing the weighted sum of orthogonal polynomials:
vals = orthopy.c1.clenshaw(a, alpha, beta, t)
orthopy is available from the Python Package Index, so use
pip install orthopy
to install.
To run the tests, simply check out this repository and run
pytest
- Robert C. Kirby, Singularity-free evaluation of collapsed-coordinate orthogonal polynomials, ACM Transactions on Mathematical Software (TOMS), Volume 37, Issue 1, January 2010
- Abedallah Rababah, Recurrence Relations for Orthogonal Polynomials on Triangular Domains, MDPI Mathematics 2016, 4(2)
- Yuan Xu, Orthogonal polynomials of several variables, arxiv.org, January 2017
This software is published under the GPLv3 license.