COVID-Twitter-BERT (CT-BERT) is a transformer-based model pretrained on a large corpus of Twitter messages on the topic of COVID-19. The v2 model is trained on 97M tweets (1.2B training examples).
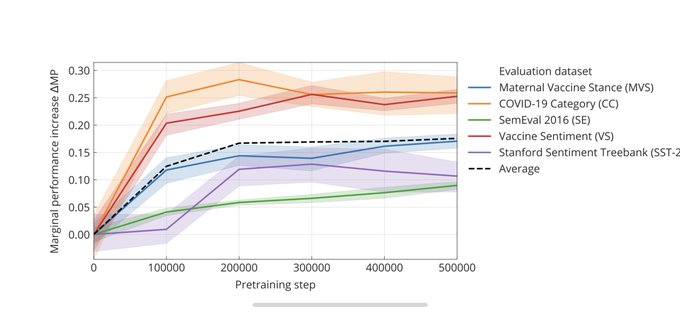
When used on domain specific datasets our evaluation shows that this model will get a marginal performance increase of 10–30% compared to the standard BERT-Large-model. Most improvements are shown on COVID-19 related and on Twitter-like messages.
This repository contains all code and references to models and datasets used in our paper as well as notebooks to finetune CT-BERT on your own datasets. If you end up using our work, please cite it:
Martin Müller, Marcel Salathé, and Per E Kummervold.
COVID-Twitter-BERT: A Natural Language Processing Model to Analyse COVID-19 Content on Twitter.
arXiv preprint arXiv:2005.07503 (2020).
For a demo on how to train a classifier on top of CT-BERT, please take a look at this Colab. It finetunes a model on the SST-2 dataset. It can also easily be modified for finetuning on your own data.
Using Huggingface (on GPU)
Using Tensorflow 2.2 (on TPUs) -
If you are familiar with finetuning transformer models, the CT-BERT-model is available both as an downloadable archive, in TFHub and as a module in Huggingface.
| Version | Base model | Language | TF2 | Huggingface | TFHub |
|---|---|---|---|---|---|
| v1 | BERT-large-uncased-WWM | en | TF2 Checkpoint | Huggingface | TFHub |
| v2 | BERT-large-uncased-WWM | en | TF2 Checkpoint | Huggingface | TFHub |
You can load the pretrained model from huggingface:
from transformers import BertForPreTraining
model = BertForPreTraining.from_pretrained('digitalepidemiologylab/covid-twitter-bert-v2')You can predict tokens using the built-in pipelines:
from transformers import pipeline
import json
pipe = pipeline(task='fill-mask', model='digitalepidemiologylab/covid-twitter-bert-v2')
out = pipe(f"In places with a lot of people, it's a good idea to wear a {pipe.tokenizer.mask_token}")
print(json.dumps(out, indent=4))
[
{
"sequence": "[CLS] in places with a lot of people, it's a good idea to wear a mask [SEP]",
"score": 0.9998226761817932,
"token": 7308,
"token_str": "mask"
},
...
]import tensorflow_hub as hub
max_seq_length = 96 # Your choice here.
input_word_ids = tf.keras.layers.Input(
shape=(max_seq_length,),
dtype=tf.int32,
name="input_word_ids")
input_mask = tf.keras.layers.Input(
shape=(max_seq_length,),
dtype=tf.int32,
name="input_mask")
input_type_ids = tf.keras.layers.Input(
shape=(max_seq_length,),
dtype=tf.int32,
name="input_type_ids")
bert_layer = hub.KerasLayer("https://tfhub.dev/digitalepidemiologylab/covid-twitter-bert/1", trainable=True)
pooled_output, sequence_output = bert_layer([input_word_ids, input_mask, input_type_ids])The script run_finetune.py can be used for training a classifier. This code depends on the official tensorflow/models implementation of BERT under tensorflow 2.2/Keras.
In order to use our code you need to set up:
- A Google Cloud bucket
- A Google Cloud VM running Tensorflow 2.2
- A TPU in the same zone as the VM also running Tensorflow 2.2
If you are a researcher you may apply for access to TPUs and/or Google Cloud credits.
Clone the repository recursively
git clone https://github.com/digitalepidemiologylab/covid-twitter-bert.git --recursive && cd covid-twitter-bertOur code was developed using tf-nightly but we made it backwards compatible to run with tensorflow 2.2. We recommend using Anaconda to manage the Python version:
conda create -n covid-twitter-bert python=3.8
conda activate covid-twitter-bertInstall dependencies
pip install -r requirements.txtSplit your data into a training set train.tsv and a validation set dev.tsv with the following format:
id label text
1224380447930683394 label_a Example text 1
1224380447930683394 label_a Example text 2
1220843980633661443 label_b Example text 3
Place these files into the folder data/finetune/originals/<dataset_name>/(train|dev).tsv (using your own dataset_name).
You can then run
cd preprocess
python create_finetune_data.py \
--run_prefix test_run \
--finetune_datasets <dataset_name> \
--model_class bert_large_uncased_wwm \
--max_seq_length 96 \
--asciify_emojis \
--username_filler twitteruser \
--url_filler twitterurl \
--replace_multiple_usernames \
--replace_multiple_urls \
--remove_unicode_symbolsThis will generate TF record files in data/finetune/run_2020-05-19_14-14-53_517063_test_run/<dataset_name>/tfrecords.
You can now upload the data to your bucket:
cd data
gsutil -m rsync -r finetune/ gs://<bucket_name>/covid-bert/finetune/finetune_data/You can now finetune CT-BERT on this data using the following command
RUN_PREFIX=testrun # Name your run
BUCKET_NAME= # Fill in your buckets name here (without the gs:// prefix)
TPU_IP=XX.XX.XXX.X # Fill in your TPUs IP here
FINETUNE_DATASET=<dataset_name> # Your dataset name
FINETUNE_DATA=<dataset_run> # Fill in dataset run name (e.g. run_2020-05-19_14-14-53_517063_test_run)
MODEL_CLASS=covid-twitter-bert
TRAIN_BATCH_SIZE=32
EVAL_BATCH_SIZE=8
LR=2e-5
NUM_EPOCHS=1
python run_finetune.py \
--run_prefix $RUN_PREFIX \
--bucket_name $BUCKET_NAME \
--tpu_ip $TPU_IP \
--model_class $MODEL_CLASS \
--finetune_data ${FINETUNE_DATA}/${FINETUNE_DATASET} \
--train_batch_size $TRAIN_BATCH_SIZE \
--eval_batch_size $EVAL_BATCH_SIZE \
--num_epochs $NUM_EPOCHS \
--learning_rate $LRTraining logs, run configs, etc are then stored to gs://<bucket_name>/covid-bert/finetune/runs/run_2020-04-29_21-20-52_656110_<run_prefix>/. Among tensorflow logs you will find a file called run_logs.json containing all relevant training information
{
"created_at": "2020-04-29 20:58:23",
"run_name": "run_2020-04-29_20-51-10_405727_test_run",
"final_loss": 0.19747886061668396,
"max_seq_length": 96,
"num_train_steps": 210,
"eval_steps": 103,
"steps_per_epoch": 42,
"training_time_min": 6.77958079179128,
"f1_macro": 0.7216383309465823,
"scores_by_label": {
...
},
...
}
Run the script 'sync_bucket_data.py' from your local computer to download all the training logs to data/<bucket_name>/covid-bert/finetune/<run_names>
python sync_bucket_data.py --bucket_name <bucket_name>In our preliminary study we have evaluated our model on five different classification datasets
| Dataset name | Num classes | Reference |
|---|---|---|
| COVID Category (CC) | 2 | Read more |
| Vaccine Sentiment (VS) | 3 | See ➡️ |
| Maternal vaccine Sentiment (MVS) | 4 | [not yet public] |
| Stanford Sentiment Treebank 2 (SST-2) | 2 | See ➡️ |
| Twitter Sentiment SemEval (SE) | 3 | See ➡️ |
If you end up using these datasets, please make sure to properly cite them.
A documentation of how we created CT-BERT can be found here.
You can cite our preprint:
@article{muller2020covid,
title={COVID-Twitter-BERT: A Natural Language Processing Model to Analyse COVID-19 Content on Twitter},
author={M{\"u}ller, Martin and Salath{\'e}, Marcel and Kummervold, Per E},
journal={arXiv preprint arXiv:2005.07503},
year={2020}
}or
Martin Müller, Marcel Salathé, and Per E. Kummervold.
COVID-Twitter-BERT: A Natural Language Processing Model to Analyse COVID-19 Content on Twitter.
arXiv preprint arXiv:2005.07503 (2020).
- Thanks to Aksel Kummervold for creating the COVID-Twitter-Bert logo.
- The model have been trained using resources made available by TPU Research Cloud (TRC) and Google Cloud COVID-19 research credits.
- The model was trained as a collaboration between Martin Müller, Marcel Salathé and Per Egil Kummervold.
- PK received funding from the European Commission for the call H2020-MSCA-IF-2017 and the funding scheme MSCA-IF-EF-ST for the VACMA project (grant agreement ID: 797876).
- MM and MS received funding through the Versatile Emerging infectious disease Observatory grant as a part of the European Commissions Horizon 2020 framework programme (grant agreement ID: 874735).
- The research was supported with Cloud TPUs from Google’s TPU Research Cloud and Google Cloud Credits in the context of COVID-19-related research”
- Martin Müller (martin.muller@epfl.ch)
- Per Egil Kummervold (per@capia.no)