Here I'm exploring the GTEx RNA-seq data which is readily available at the GTEx portal. This small R project will generate an R data frame for easy exploration (currently working on making it a tidy data set). Below I have a couple of UMAP plots on the samples, for exploratory/exemplary analyses.
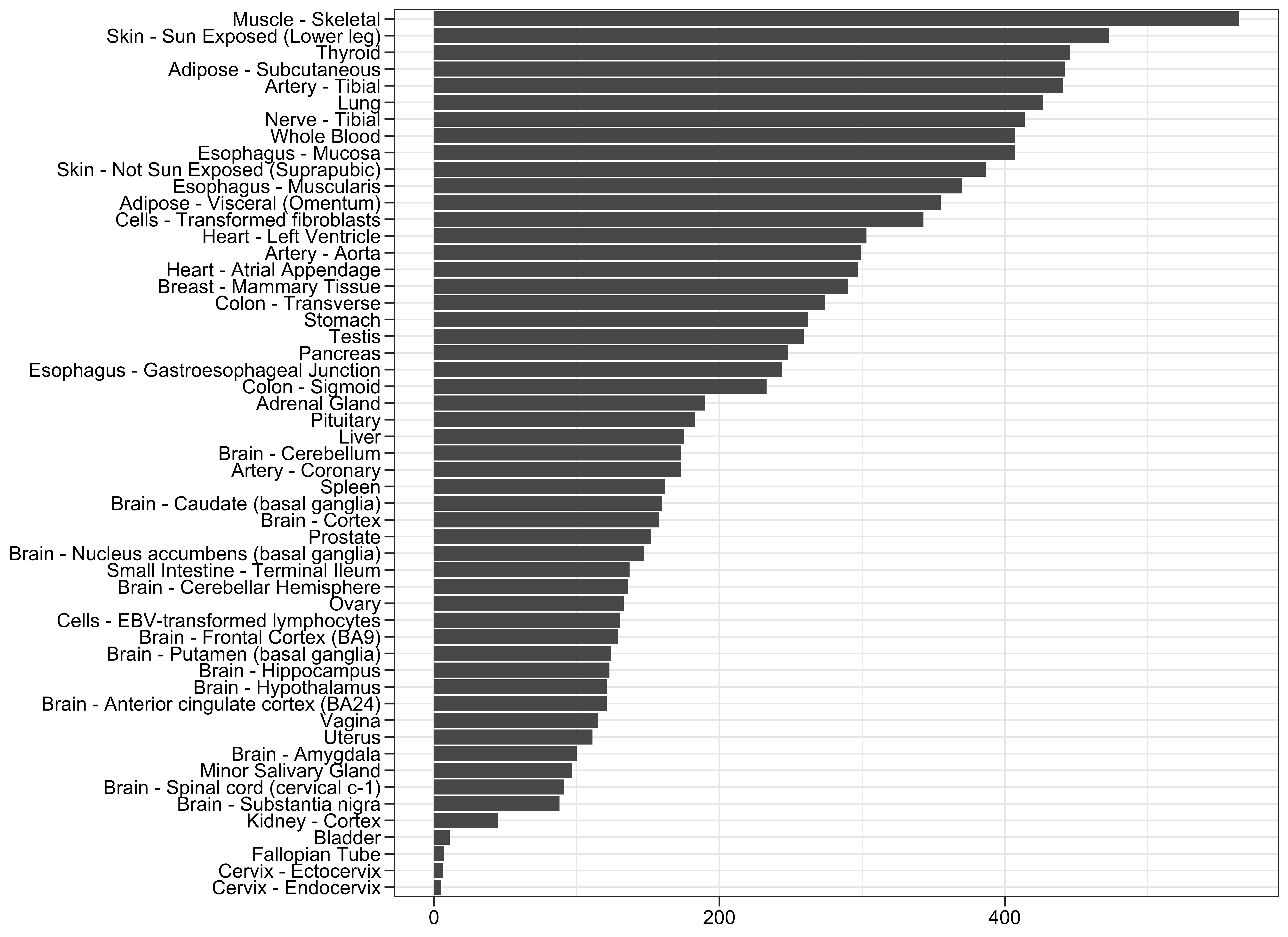
One of the aspects of interest is to check how many sub-tissues were sampled and made available. From the data set:
samples <- samples %>%
dplyr::mutate(., tissue = sample_annotations$SMTSD[match(X1, sample_annotations$SAMPID)]) %>%
dplyr::mutate(., type = sample_annotations$SMAFRZE[match(X1, sample_annotations$SAMPID)]) %>%
dplyr::mutate(., broader_type = sample_annotations$SMTS[match(X1, sample_annotations$SAMPID)])
samples %>%
dplyr::group_by(., tissue) %>%
dplyr::count() %>%
ggplot() +
geom_col(aes(y = n, x = forcats::fct_reorder(tissue, n))) +
coord_flip() +
theme_bw() +
theme(axis.text = element_text(color = "black", size = 12),
axis.title = element_blank(),
axis.ticks.length = unit(0.2, "cm"))which will produce:
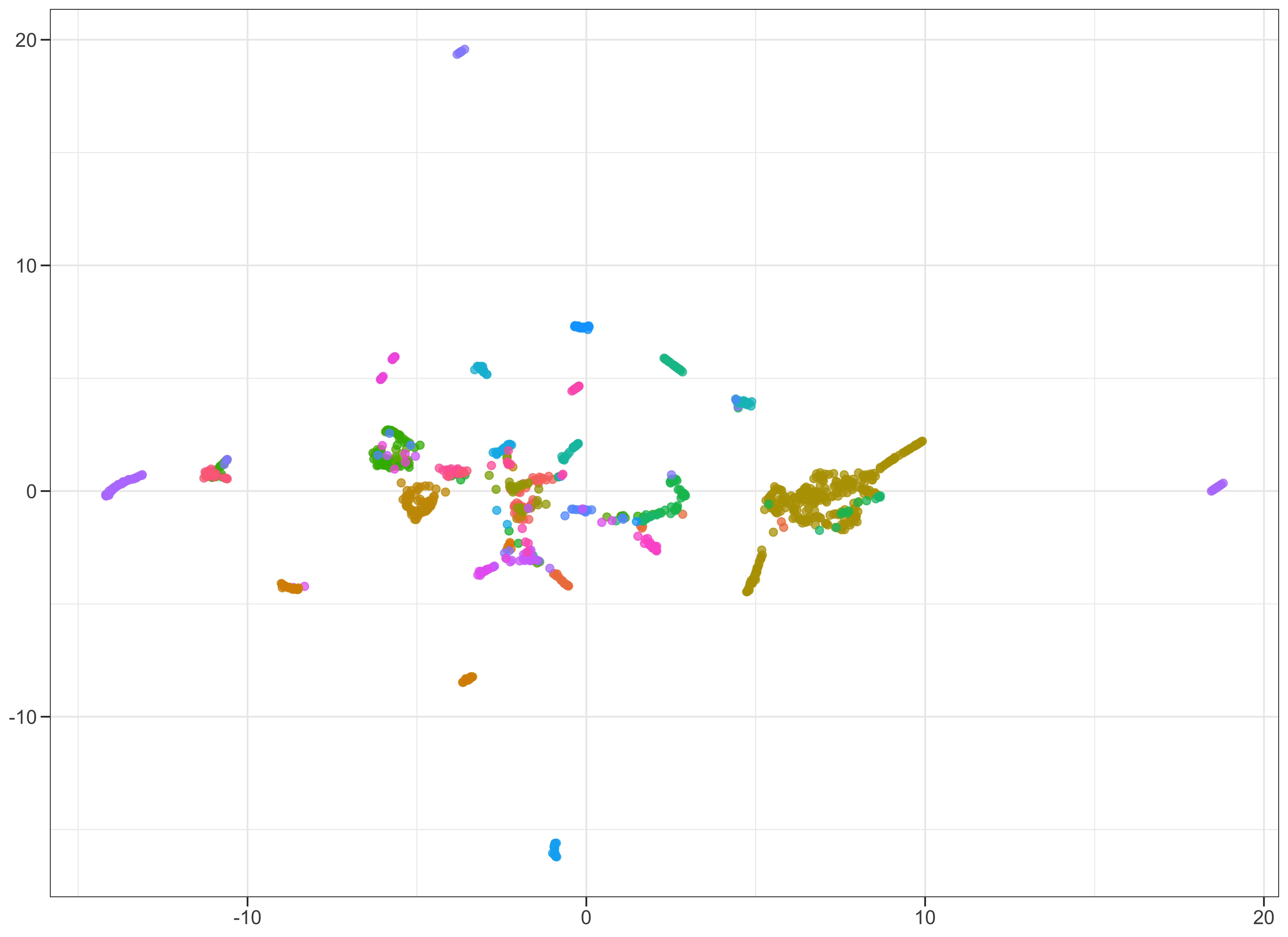
Running UMAP on a subset of samples and subset of genes. In R:
umap_plot <- umap::umap(d = t(data_mat),
method = "naive",
random_state = 123,
n_epochs = 100,
min_dist = 0.1,
n_neighbors = 15,
metric = "cosine")which produces:
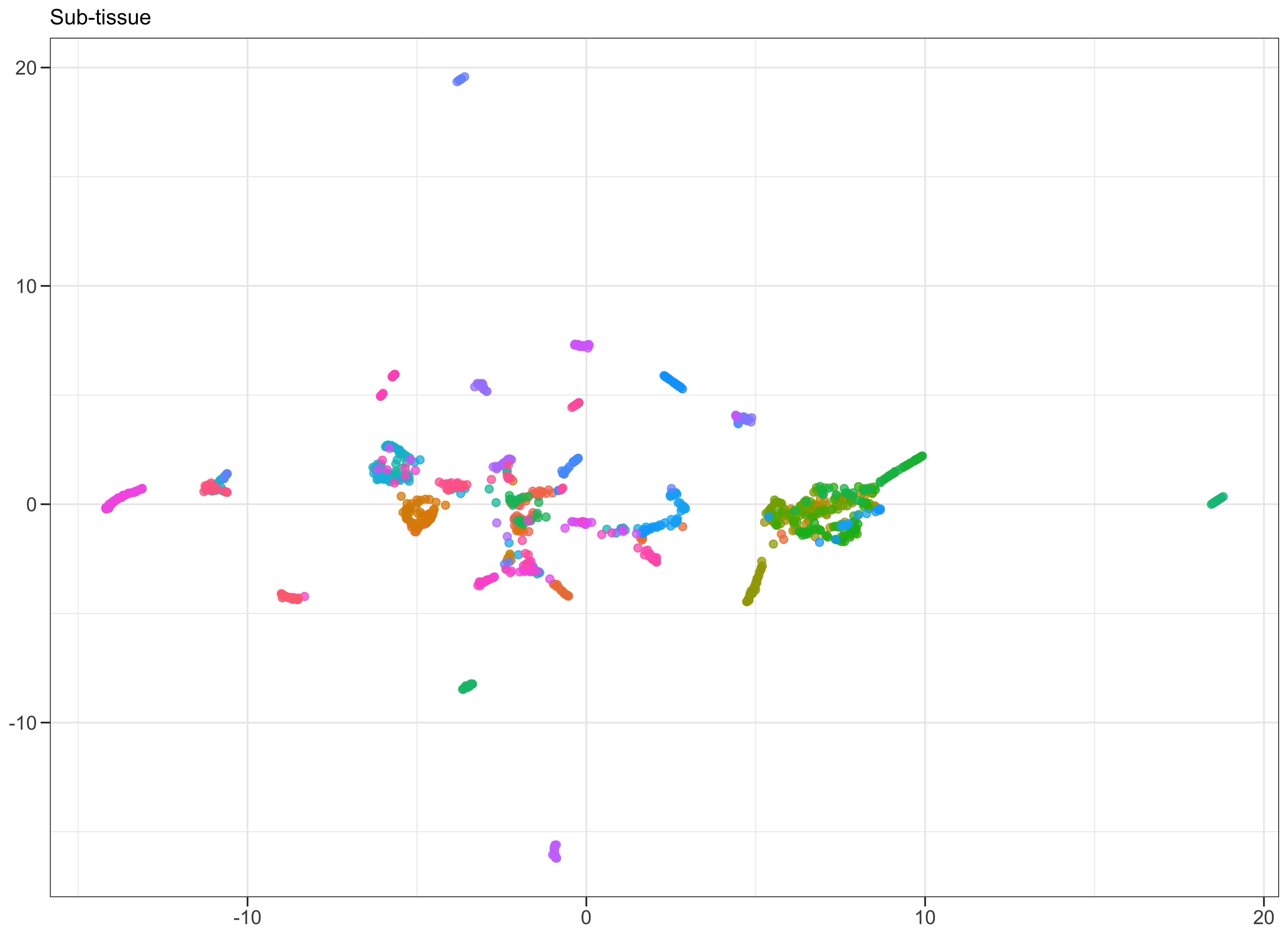
Same as before. Plot: