- Overview
- Machine Learning in Bioinformatics
- Distance Functions
- Hierarchical Clustering
- K-Means Clustering
- Summary
It's a cozy friday evening and your girlfriend/boyfriend comes over to your house to watch netflix and chill. As you log in, you are swarmed with advertisements recommending you to watch "The Avengers", "Superman", and "James Bond". The suggestions all sound good, so how did Netflix know that you would like these movies? Netflix uses a special type of classification algorithm that grouped you with the action movie enthusiasts. They noticed which movies you watched in the past and predicted what type of movies you would like in the future based on those past decisions. Making predictions and conclusions based on data is the crux of machine learning.
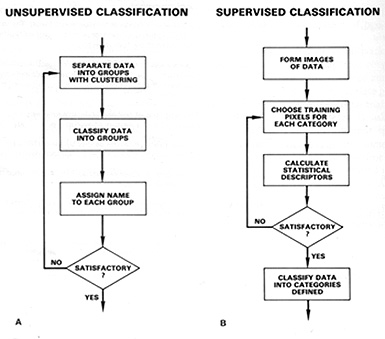
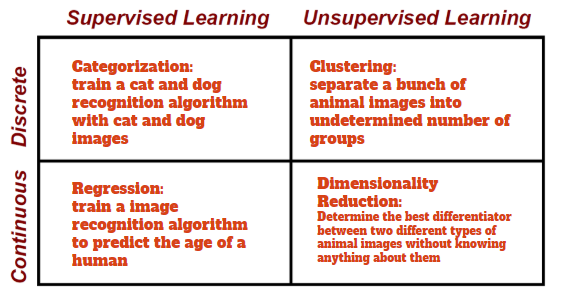
Machine learning refers to the general type of algorithm that makes assertions or predictions based on data. Machine learning can be used to categorize individuals into certain groups based on shared similarities or recognize certain patterns that match an individual. There are many different kinds of questions that can be answered with machine learning, hence it is split into several domains: supervised vs unsupervised, clustering vs categorization, and continuous vs discrete.
An algorithm is said to be supervised if the potential types, or labels of the data are known. On the other hand, an unsupervised algorithm does not have output labels and can work with anonymous data. Unsupervised algorithms are used when the grouping of the data is more important than the label themselves, and supervised algorithms are used when the type of the group provides more meaning.
The second domain of machine learning is continuous vs discrete. Continuous algorithms have outputs on continuous, or flowing, spectrum. Discrete algorithms produce information in distinct, well-defined buckets.
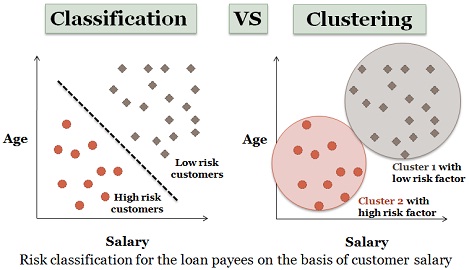
Lastly, a machine learning algorithm could be classifying or clustering. Classification and clustering algorithms often answer the same question, but differ in their implementations. In general, classification algorithms aim to find the best way to separate data in classes, whereas clustering algorithms strive to group data into cliques. Depending on the circumstance, classification and clustering may give different results based on the input data.
There are many applications of machine learning in bioinformatics. Machine learning algorithms are used in image classification, detecting variation in rare diseases, and computing phylogenetic trees. In biomedical informatics, we see machine learning used in precision medicine. The general idea of precision medicine is to make disease predictions based on an individual's genomic or transcriptomic (RNA-Seq) data. We can use this personalized information to predict potential cancer-causing genes or discover subtypes of a disease.
In RNA-Seq data, a datapoint is a multidimensional vector. Each row corresponds to a gene, and each column represents an individual. Often, we use clustering algorithms to group individuals together who have the same variation of a disease to diagnose the best therapy.
In the subsequent sections, we will examine two commonly used clustering algorithms used in bioinformatics.
Before we discuss the details of clustering algorithms, we need to define a distance function. The distance function is a function that computes the difference between two points by some mathematical quantity. The most common distance function uses Euclidean distance, or the distance between points in geometric space. Another distance function could be Hamming distance, the number of different nucleotides in DNA strings.
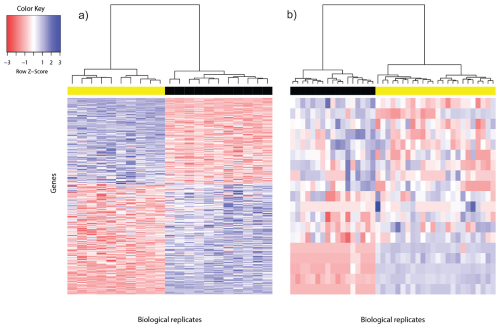
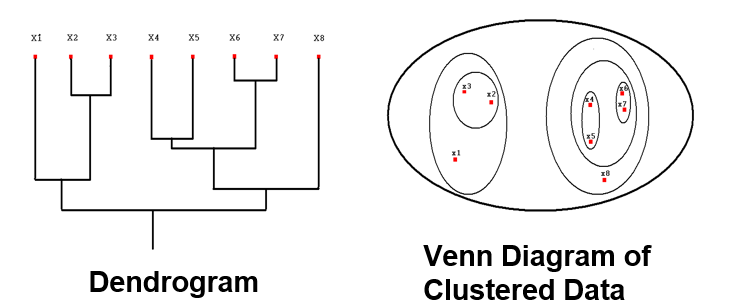
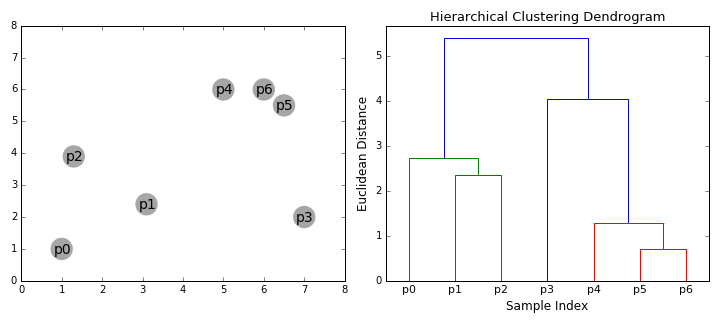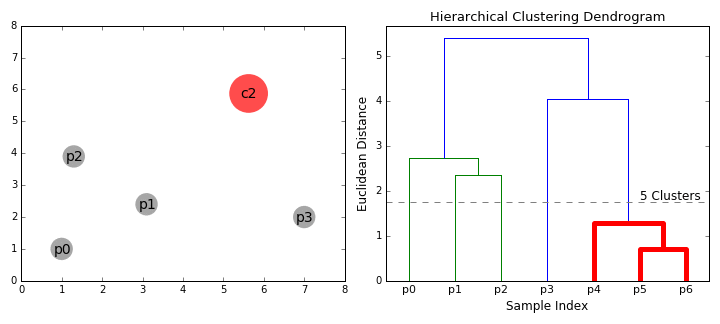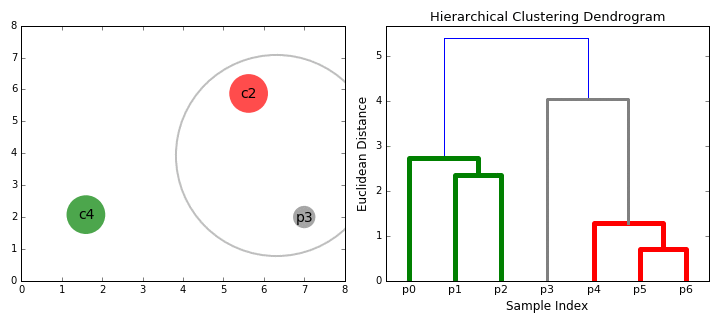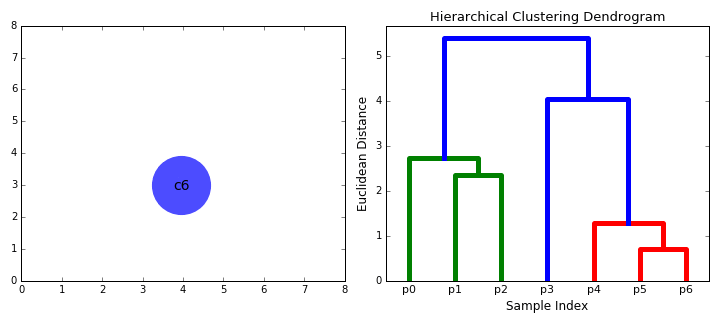
In hierarchical clustering, we compute a dendrogram that separates data points. A dendrogram is a diagram that illustrates the arrangement of clusters in a tree. However, it could also be visualized in a heatmap or venn diagram. In the above RNA-Seq example, we see that a dendrogram is produced above the heatmap that clusters individuals together based on similarity.
The following is the general procedure of a hierarchical clustering algorithm:
- Calculate the similarity (distance function) between all possible combinations of two profiles
- Place each profile in a separate cluster.
- Group the two most similar clusters together to form a new cluster.
- Recalculate the similarity between the new cluster and all the remaining clusters.
- Repeat steps 3 and 4 until all of the profiles end up in one large cluster.
In hierarchical clustering, you have several clustering methods that each use a different distance function:
- Unweighted Pair Group Method (UPGMA) - Calculates the average distance from each point in the cluster to all other points in another cluster
- Single Linkage - Measures dissimilarity between two clusters as the minimum dissimilarity between members of the two clusters
- Complete Linkage - Measures dissimilarity between two clusters as the greatest dissimilarity between members of the two clusters
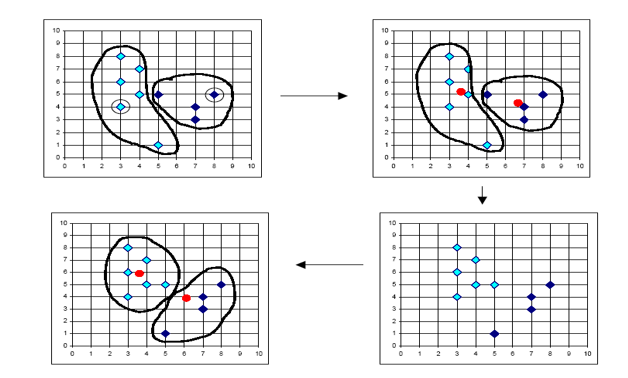
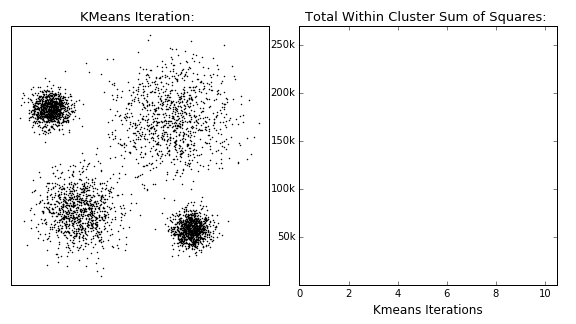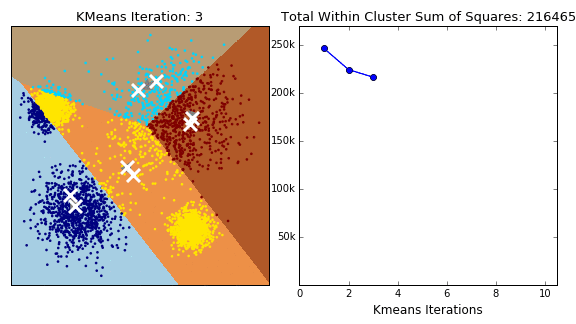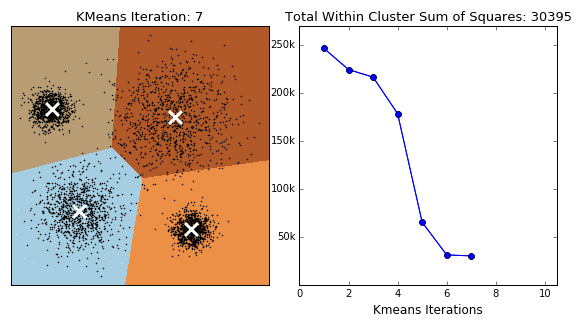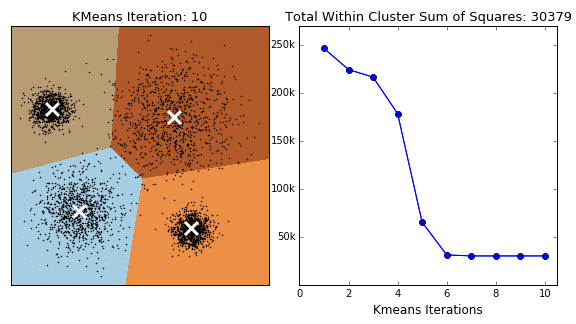
In K-Means clustering, we compute groups by minimizing the distance of each point to to its group's mean. Unlike hierarchical clustering, k-means clustering must arbitrarily choose k, the number of clusters, and consequently select k points to serve as the initial means for each cluster. At each iteration, we continuously reassign points such that they are grouped with the clusters that minimize the point's distance to the cluster's mean.
The following is the general procedure of the k-means clustering algorithm:
- Select the number of clusters K
- Select the K starting points to serve as the initial cluster means
- Iterate through each point and calculate the distance between the datum and each cluster's mean
- Assign the datum to the cluster whose mean is closest to that point
- Repeat steps 3 and 4 until convergence - when points are no longer reassigned
Note that K-Means clustering does not always guarantee termination. An upper bound for the number of iterations should be assigned to prevent infinite loops. Additionally, the selection of the initial points can change the outcome. Perhaps the selection of the initial points should be given careful consideration rather than an arbitrary choosing.
Machine Learning is perhaps the greatest export of computing to biology. Any algorithm that classifies or categorizes data utilizes some form of machine learning. In computer science, we see machine learning in image classification and recommender systems. In bioinformatics, machine learning tackles important medical problems such as categorizing patients into subtypes of a disease or detecting genes responsible for pathology. In the future, machine learning will become an essential component of bioinformatics as we aggregate more and more biological data. Machine learning helps us deal with the information explosion of the 21st century and will lay the foundation for precision medicine and gene-function discovery.
- Sheng Zhong BENG 183
- Victoria Tom, Joey Sun BENG 183
- https://dashee87.github.io/data%20science/general/Clustering-with-Scikit-with-GIFs/