A wrapper to estimate admixture proportions using qpAdm implemented in AdmixTools
qpAdm-wrapper has two modes:
- write: to generate input files for qpAdm analysis that included all combinations of target, source and right populations and list all results
- read: to read all log files produced by qpAdm and evaluate them based on p-value and Z scores.
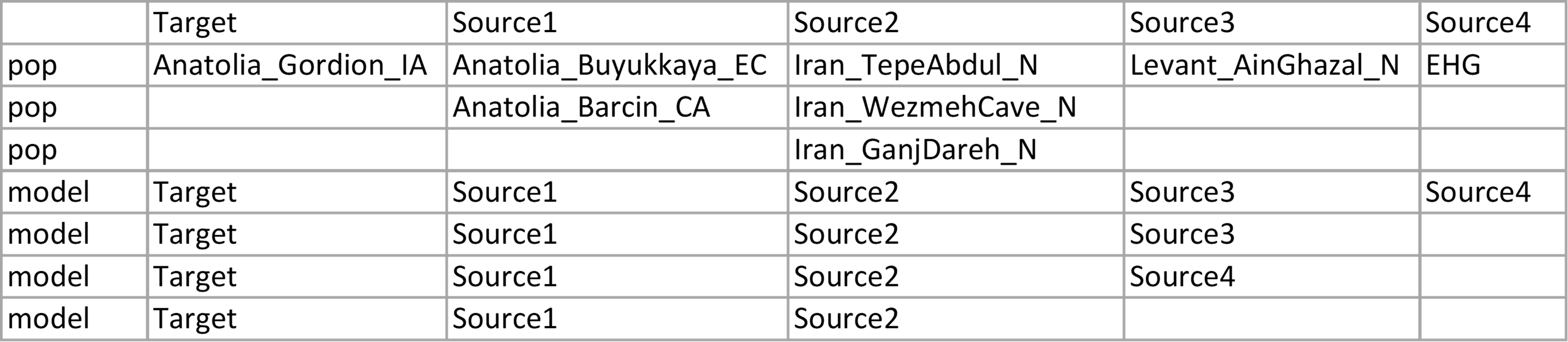
Assume we have a target population (Anatolia_Gordion_IA) that we want to estimate admixture populations by using the different numbers of sources (2,3 or 4) with various possible ancestry sources from the same gene pool (i.e. to explain Anatolian-related ancestry in the target population "Anatolia_Gordion_IA", we want to use either Anatolia_Buyukkaya_EC or Anatolia_Barcin_CA) as below. To run all possible combinations of target and source populations by using Admixtools, we have to create 23 different left files and 23 different par files. When we want to model more than one target population the number of required input files will increase.
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_TepeAbdul_N
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_WezmehCave_N
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_GanjDareh_N
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_TepeAbdul_N
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_WezmehCave_N
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_GanjDareh_N
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_TepeAbdul_N EHG
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_WezmehCave_N EHG
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_GanjDareh_N EHG
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_TepeAbdul_N EHG
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_WezmehCave_N EHG
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_GanjDareh_N EHG
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_TepeAbdul_N Levant_AinGhazal_N
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_WezmehCave_N Levant_AinGhazal_N
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_GanjDareh_N Levant_AinGhazal_N
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_TepeAbdul_N Levant_AinGhazal_N
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_WezmehCave_N Levant_AinGhazal_N
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_GanjDareh_N Levant_AinGhazal_N
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_TepeAbdul_N Levant_AinGhazal_N EHG
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_WezmehCave_N Levant_AinGhazal_N EHG
Anatolia_Gordion_IA Anatolia_Buyukkaya_EC Iran_GanjDareh_N Levant_AinGhazal_N EHG
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_TepeAbdul_N Levant_AinGhazal_N EHG
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_WezmehCave_N Levant_AinGhazal_N EHG
Anatolia_Gordion_IA Anatolia_Barcin_CA Iran_GanjDareh_N Levant_AinGhazal_N EHG
qpAdm-wrapper can prepare all these input files and produce a script file (either in sbatch or bash format) to run multiple qpAdm models using multiple threads from a simple input file (info file) like below.
This info file should be prepared as a data frame that has two indexes called 'pop' and 'model'. The rows containing information about 'left' populations are indexed as 'pop', while the rows containing the model are indexed as 'model'. The header of dataframe (column names) indicates target and possible ancestry sources. Column names can be changed, but the first column should always contain target population(s).
The info file should be tab-separeted and can contain empty cells. However, empty cells could be filled by NA's to avoid confusion.
You could give Admixtools bin path by using --admixtools_path option.
- numpy
- pandas
- argparse
- os
- itertools
- base par file (see example par file)
- population info file (see example info file)
-i --input <filename> "Input file contains information about target and sources populations and models"
-o --outgroup_name <filename> "The file contains a list of the right populations (1 per line) "
-p --par <filename> "base par file"
-w --write <BOOL> "write mode [default: True], when read mode is True [default: False]"
-r --read <BOOL> "read mode [default: False]"
--run_mode <STR> "Output script file could be either in sbatch or bash format [default: sbatch]"
--admixtools_path <STR> "path of the Admixtools bin"
-t --threads <INT> "number of threads"
-ho --host_name <STR> "if the run_mode is sbatch, partition/queue in which to run the job"
--out_path <STR> "the directory where output files are written"./qpAdm-wrapper.py -i gordion -p par -o Base12_CHG -ho chimp -t 80 The header of sbatch output file will be like below:
#!/bin/bash -l
#SBATCH -J qpAdm
#SBATCH -p chimp #can be specified by -ho option
#SBATCH -n 80 #can be specified by -t option
#SBATCH -t 5-00:00:00
#SBATCH -D ./ #can be specified by --out_path option
#SBATCH -o slurm-%j-%N-%u.out
#SBATCH -e slurm-%J-%N-%u.err
export PATH=$PATH:/usr/local/sw/AdmixTools-7.0.2/bin/ #can be specified by --admixtools_path option
If your cluster uses different slurm configurations or specifies options differently, you can modify the create_sbatch function in the script. Or you should modify the sbatch output file manually. You can also create bash file as output and define options during submitting. For more detail you can check slurm-sbatch website.
./qpAdm-wrapper.py -i gordion -p par -o Base12_CHG --run_mode bash -t 80 To summarize results from all log files after qpAdm runs have finished
./qpAdm-wrapper.py -i gordion -p par -o Base12_CHG -r TrueThis create the output file named results_poplist_gordion.txt
You can copy and paste results to the excel file in examples/output_files/qpAdm_results_template.xltx, and easily compare all results by checking feasible and z_eval columns.