This repo contains supporting code for "Preference Optimization for Molecular Language Models".
We first recommend setting up a conda environment for this project. This can be done with
conda create -n pref_opt_for_mols python=3.9Once created, we can activate the environment and install the required dependencies:
conda activate pref_opt_for_mols
pip install -r requirements.txtNow the package can be installed from source from this directory by running
pip install .Before pre-training on the MOSES benchmark set, we need to download the MOSES training and testing sets of SMILES, and place them in scripts/data/train.csv and scripts/data/test.csv.
Once these are downloaded, we can run pre-training on either the GPT or CharRNN models using the scripts/pretrain.py script. This requires a config file be defined first. Examples for both models can be found in scripts/configs/pretrain/.
For example, to train the GPT model we can run the following (from the scripts/ directory):
python pretrain.py --arch gpt --config configs/pretrain/gpt_demo.json --device 0Similarly, to train the CharRNN model we can run
python pretrain.py --arch rnn --config configs/pretrain/rnn_demo.json --device 0After pre-training, molecules can be sampled using the scripts/sample.py script, e.g. using
python sample.py --arch gpt --model_path checkpoints/smiles-gpt-demo/ --num_batches 2 --batch_size 512 --out demo_smiles.csv --device 0which will sample 2 batches of 512, and write them to a file demo_smiles.csv.
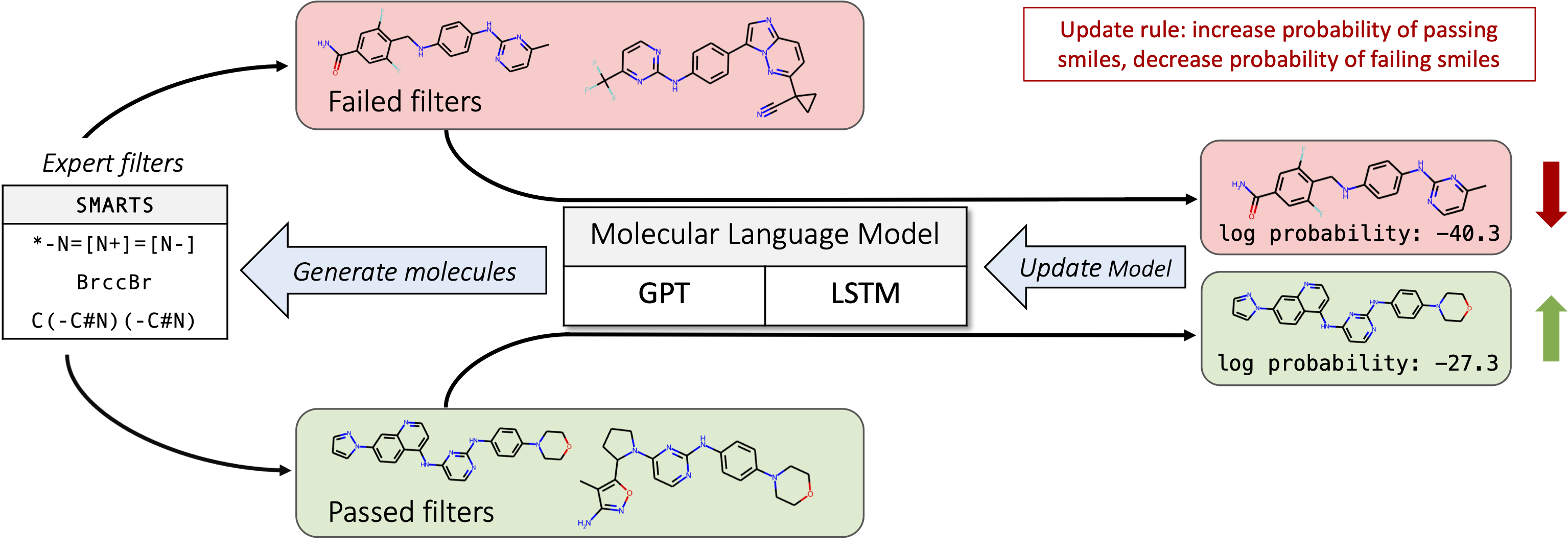
After pre-training, models can be fine-tuned with DPO. This is a multi-step process that requires molecules to be sampled, filtered/scored, and used for preference optimization.
Sampling can be done with scripts/sample.py as mentioned previously. To filter the sampled molecules for positive/negative examples, use the scripts/filter.py script like so
python filter.py --smiles sampled_smiles.csv --out sampled_smiles_filtered.csv --method mcf --batch_size 128This will add a column label to the sampled smiles dataframe and write the updated data to sampled_smiles_filtered.csv. The method controls the objective we're trying to fine-tune. Currently, only mcf is supported (optimize for common medchem checks, such as # of chiral centers and SMARTS filters).
Next, train on the filtered set with scripts/finetune.py, e.g.
python finetune.py --config configs/dpo/demo.json --name DemoExperiment --device 0which will train/save a DPO-optimized model according to the hyperparameters set in demo.json. We recommend using the same pre-trained base model for sampling and fine-tuning.