# If the following error occurs:
# Error in utils::download.file(url, path, method = method, quiet = quiet, :
# download from 'https://api.github.com/repos/Asa12138/ReporterScore/tarball/HEAD' failed
# options(timeout = 600)
devtools::install_github("dongdongdong7/MetaboRich")The input should be a data.frame or tibble with four columns, as follows:
library(MetaboRich)
data("example_data", package = "MetaboRich")> dplyr::as_tibble(example_data)
# A tibble: 168 × 4
id logfc pvalue qvalue
<chr> <dbl> <dbl> <dbl>
1 C00082 1.76 0.00000289 0.000599
2 C12512 -2.50 0.0000323 0.00334
3 C16300 -1.71 0.000215 0.0148
4 C04886 0.807 0.000438 0.0187
5 C00486 0.807 0.000450 0.0187
6 C01996 0.623 0.000623 0.0215
7 C03413 0.622 0.00126 0.0291
8 C00366 -0.256 0.00198 0.0357
9 C01019 -0.811 0.00198 0.0357
10 C16618 -0.599 0.00207 0.0357
# ℹ 158 more rows
# ℹ Use `print(n = ...)` to see more rowsThere are two main types of built-in data that you can retrieve through data():
data("metabolitesList", package = "MetaboRich")
names(metabolitesList) # "hmdb" "ramp"
data("setsList", package = "MetaboRich")
names(setsList)
# [1] "kegg_pathway" "smpdb_pathway" "hmdb_disease" "markerdb_condition" "classyfire"
# [6] "reactome_pathway" "wiki_pathway" MetabolitesList contains metabolite information from two databases, HMDB and RaMP. These databases are organized into the form of tibble, called metabolites_tibble, as follows:
> metabolitesList$hmdb
# A tibble: 217,920 × 23
hmdb_id name formula mw monoisotop_mass smiles inchi inchikey foodb_id kegg_id chemspider_id chebi_id
<chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 HMDB0000001 1-Met… C7H11N… 169. 169. CN1C=… InCh… BRMWTNU… FDB0935… C01152 83153 50599
2 HMDB0000002 1,3-D… C3H10N2 74.1 74.1 NCCCN InCh… XFNJVJP… FDB0052… C00986 415 15725
3 HMDB0000005 2-Ket… C4H6O3 102. 102. CCC(=… InCh… TYEYBOS… FDB0303… C00109 57 30831
4 HMDB0000008 2-Hyd… C4H8O3 104. 104. CC[C@… InCh… AFENDNX… FDB0218… C05984 389701 50613
5 HMDB0000010 2-Met… C19H24… 300. 300. [H][C… InCh… WHEUWNK… FDB0218… C05299 389515 1189
6 HMDB0000011 3-Hyd… C4H8O3 104. 104. C[C@@… InCh… WHBMMWS… FDB0218… C01089 83181 17066
7 HMDB0000012 Deoxy… C9H12N… 228. 228. OC[C@… InCh… MXHRCPN… FDB0218… C00526 13118 16450
8 HMDB0000014 Deoxy… C9H13N… 227. 227. NC1=N… InCh… CKTSBUT… FDB0218… C00881 13117 15698
9 HMDB0000015 Corte… C21H30… 346. 346. [H][C… InCh… WHBHBVV… FDB0218… C05488 389582 28324
10 HMDB0000016 Deoxy… C21H30… 330. 330. [H][C… InCh… ZESRJSP… FDB0064… C03205 5932 16973
# ℹ 217,910 more rows
# ℹ 11 more variables: drugbank_id <chr>, phenol_explorer_compound_id <chr>, wikipedia_id <chr>,
# knapsack_id <chr>, bigg_id <chr>, metlin_id <chr>, vmh_id <chr>, cas_id <chr>, synonyms <list>,
# iupac_name <chr>, traditional_iupac <chr>
# ℹ Use `print(n = ...)` to see more rowsMetabolites_tibble can also be arranged by users, which contains three columns of main information databasename1_id, name, and databasename2_id to be used for id conversion of function id_mapping.
Several metabolite sets/function sets are currently collected in setsList. The common pathway sets are kegg_pathway, smpdb_pathway, reactome_pathway and wiki_pathway. Disease set: hmdb_disease, markerdb_condition; Structure classification set: classyfire.
> setsList$kegg_pathway
# A tibble: 289 × 5
id name type metabolites metabolites_name
<chr> <chr> <chr> <list> <list>
1 hsa00010 Glycolysis / Gluconeogenesis Metabolism; Carbohydrate me… <chr [31]> <chr [31]>
2 hsa00020 Citrate cycle (TCA cycle) Metabolism; Carbohydrate me… <chr [20]> <chr [20]>
3 hsa00030 Pentose phosphate pathway Metabolism; Carbohydrate me… <chr [37]> <chr [37]>
4 hsa00040 Pentose and glucuronate interconversions Metabolism; Carbohydrate me… <chr [59]> <chr [59]>
5 hsa00051 Fructose and mannose metabolism Metabolism; Carbohydrate me… <chr [55]> <chr [55]>
6 hsa00052 Galactose metabolism Metabolism; Carbohydrate me… <chr [46]> <chr [46]>
7 hsa00053 Ascorbate and aldarate metabolism Metabolism; Carbohydrate me… <chr [57]> <chr [57]>
8 hsa00500 Starch and sucrose metabolism Metabolism; Carbohydrate me… <chr [37]> <chr [37]>
9 hsa00520 Amino sugar and nucleotide sugar metabolism Metabolism; Carbohydrate me… <chr [118]> <chr [118]>
10 hsa00620 Pyruvate metabolism Metabolism; Carbohydrate me… <chr [32]> <chr [32]>
# ℹ 279 more rows
# ℹ Use `print(n = ...)` to see more rowsUsers can also imitate the above examples to build their own metabolite sets.
The R package contains three enrichment functions, ORA, KSTEST, and GRSA.
enrichmentRes_ora <- ora(input = example_data, enrich_tibble = setsList$kegg_pathway, enrich_type = "all", N_type = "database", adjust = "fdr", thread = 2)
enrichmentRes_kstest <- kstest(input = example_data, enrich_tibble = setsList$kegg_pathway, adjust = "fdr", thread = 2)
enrichmentRes_grsa <- grsa(input = example_data, enrich_tibble = setsList$kegg_pathway, adjust = "fdr", thread = 2)The results of enrichment are presented in the form of tibble
> library(magrittr)
> enrichmentRes_ora %>% dplyr::filter(pvalue < 0.05) %>% dplyr::arrange(pvalue)
# A tibble: 10 × 9
id name type set inset insetIDs direaction pvalue qvalue
<chr> <chr> <chr> <int> <int> <chr> <dbl> <dbl> <dbl>
1 hsa04974 Protein digestion and absorption Organismal S… 47 6 C00082;… 0.515 2.48e-5 0.00716
2 hsa05230 Central carbon metabolism in cancer Human Diseas… 37 5 C00082;… 0.611 9.48e-5 0.0137
3 hsa00970 Aminoacyl-tRNA biosynthesis Genetic Info… 52 5 C00082;… 0.451 4.89e-4 0.0471
4 hsa04917 Prolactin signaling pathway Organismal S… 11 2 C00082;… 1 8.43e-3 0.609
5 hsa00470 D-Amino acid metabolism Metabolism; … 69 4 C00739;… 0 1.17e-2 0.674
6 hsa04976 Bile secretion Organismal S… 174 6 C00486;… 0.654 2.36e-2 1
7 hsa00360 Phenylalanine metabolism Metabolism; … 49 3 C00082;… 0.841 2.49e-2 1
8 hsa04080 Neuroactive ligand-receptor interaction Environmenta… 53 3 C01996;… 0.690 3.06e-2 1
9 hsa00220 Arginine biosynthesis Metabolism; … 23 2 C00049;… 0 3.52e-2 1
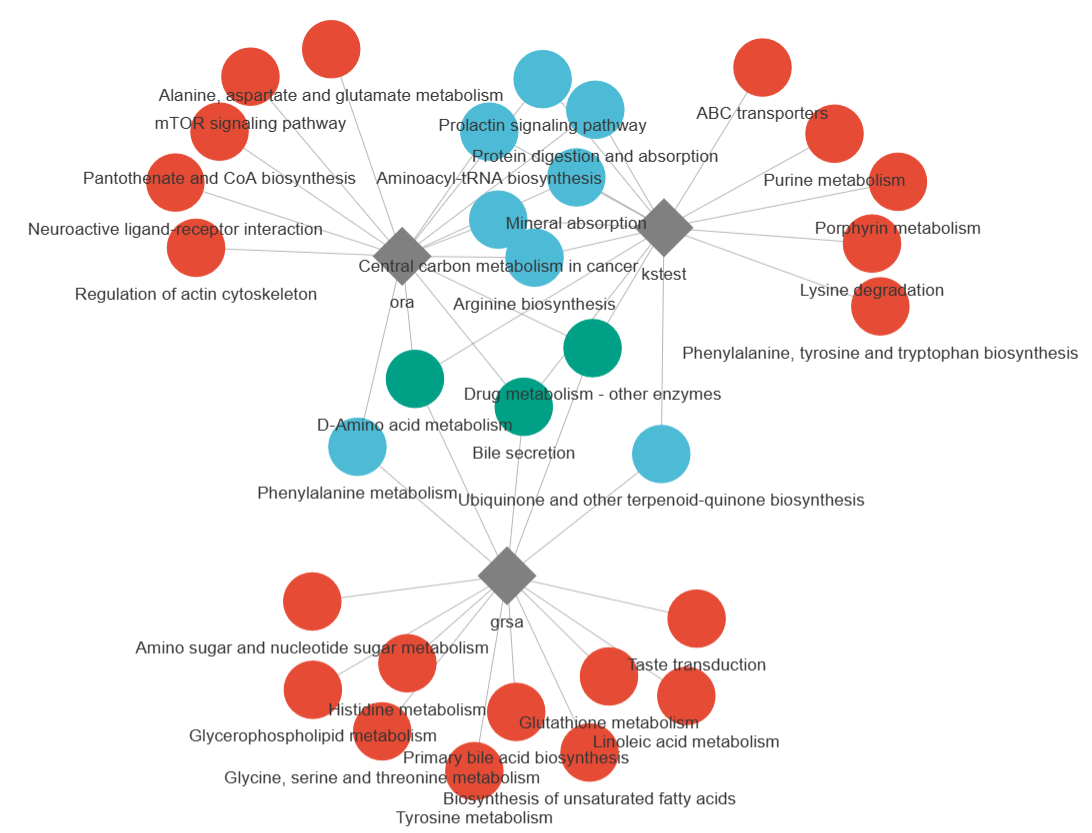
10 hsa00983 Drug metabolism - other enzymes Metabolism; … 63 3 C16618;… 0 4.74e-2 1 If the id of the metabolite in enrich_tibble is different from the id of the input, an error can occur, so you may first do an id_mapping to make sure that the id of the input is the same as that in enrich_tibble.
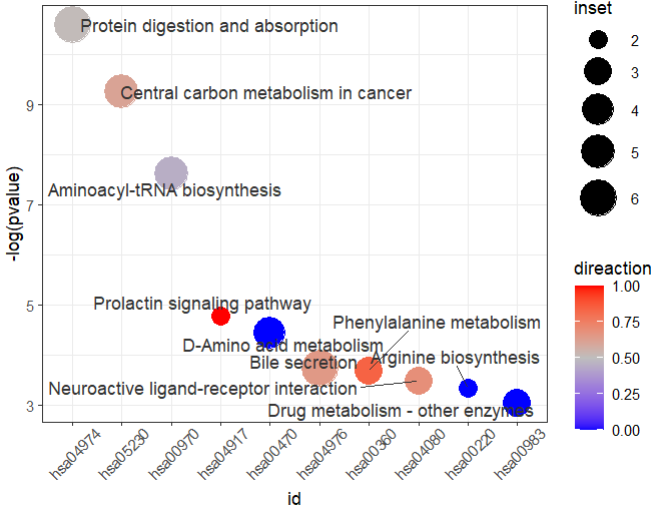
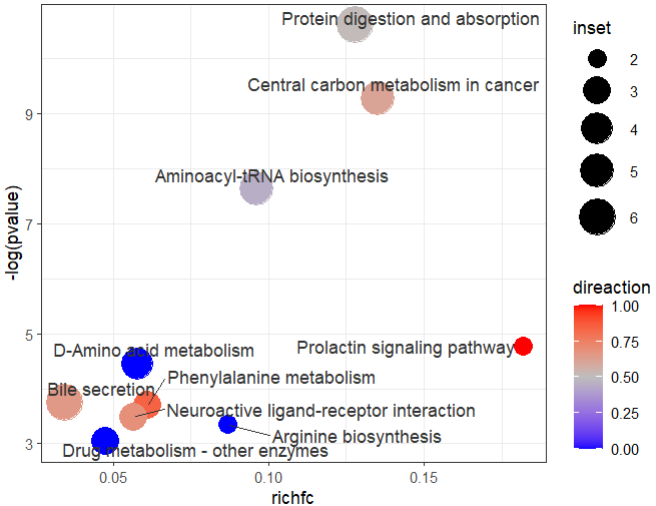
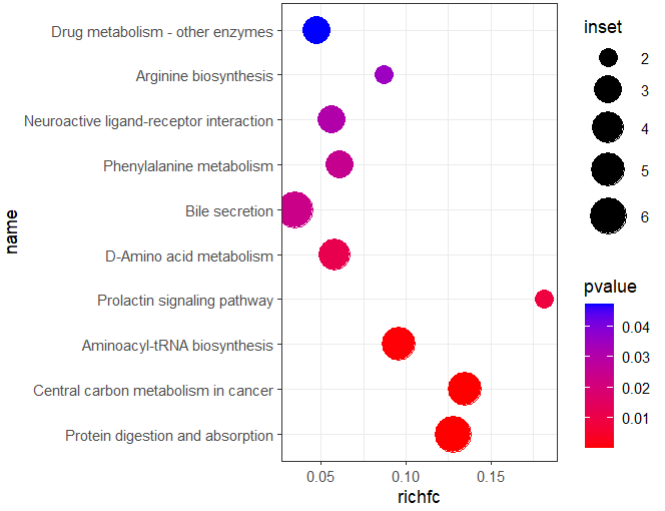
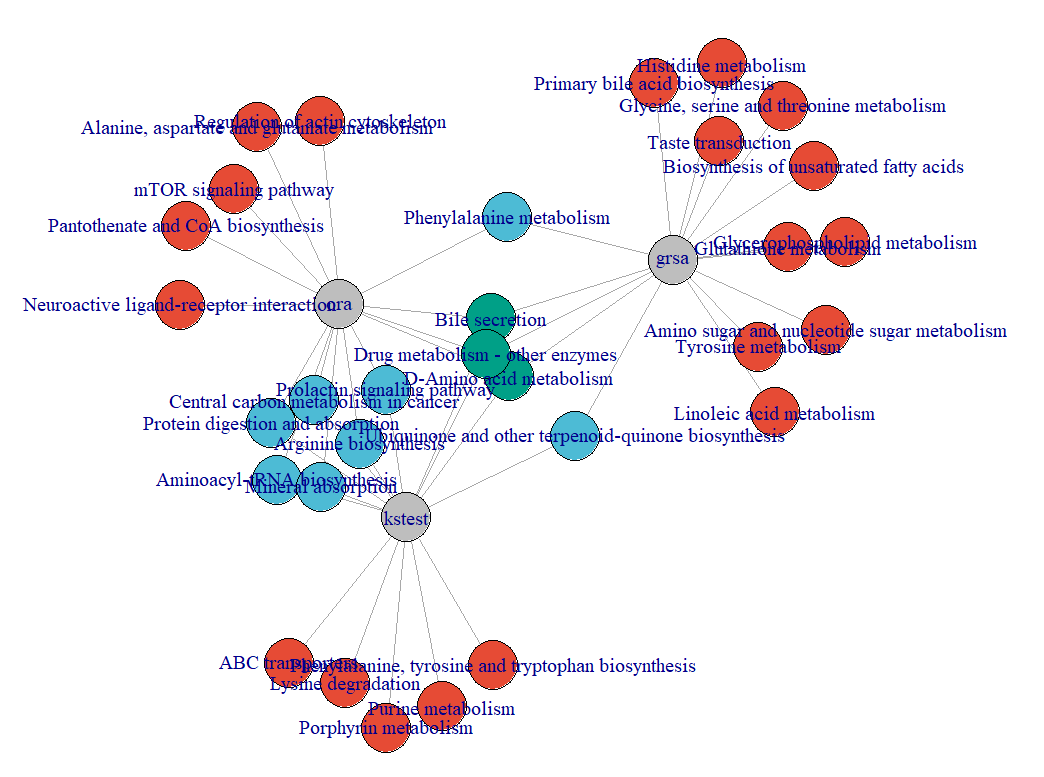
example_data <- id_mapping(input = example_data, from = "kegg_id", to = "hmdb_id", metabolites_tibble = metabolitesList$hmdb)plot_enrichmentRes(enrichmentRes = enrichmentRes_ora, plot_type = 1)plot_enrichmentRes(enrichmentRes = enrichmentRes_ora, plot_type = 2)plot_enrichmentRes(enrichmentRes = enrichmentRes_ora, plot_type = 3)plot_compareRes(enrichmentResList = list(ora = enrichmentRes_ora, kstest = enrichmentRes_kstest, grsa = enrichmentRes_grsa), plot_type = 1)plot_compareRes(enrichmentResList = list(ora = enrichmentRes_ora, kstest = enrichmentRes_kstest, grsa = enrichmentRes_grsa), plot_type = 2)