This repository contains source code for the paper [Inquire, Interact, and Integrate: A Proactive Agent Collaborative Framework for Zero-Shot Multimodal Medical Reasoning], which is still underreview.
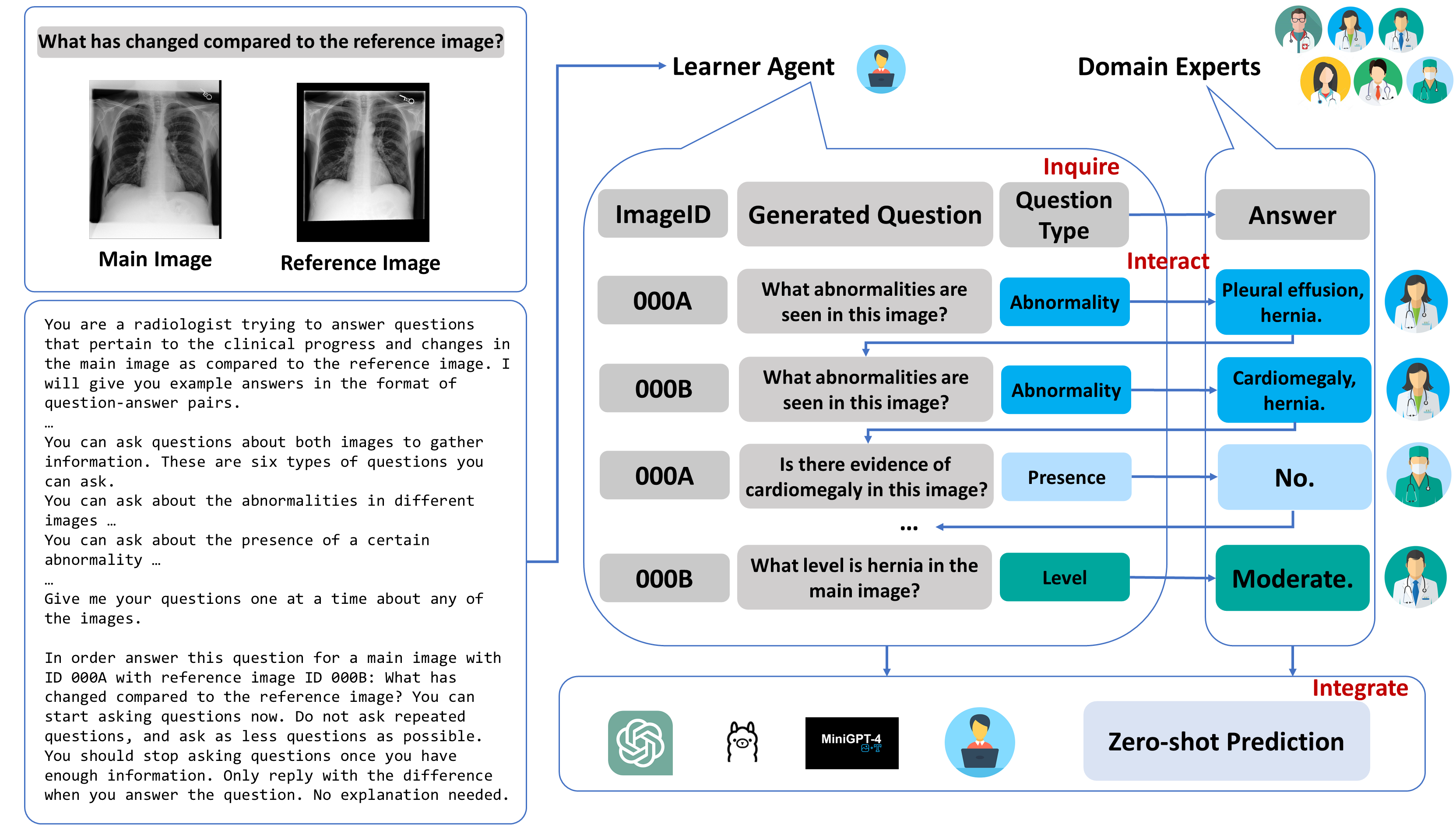
The adoption of large language models (LLMs) in healthcare has attracted significant research interest. However, their performance in healthcare remains under-investigated and potentially limited, due to i) they lack rich domain-specific knowledge and medical reasoning skills; and ii) most state-of-the-art LLMs are unimodal, text-only models that cannot directly process multimodal inputs. To this end, we propose a multimodal medical collaborative reasoning framework MultiMedRes, which incorporates a learner agent to proactively gain essential information from domain-specific expert models, to solve medical multimodal reasoning problems. Our method includes three steps: i) Inquire: The learner agent first decomposes given complex medical reasoning problems into multiple domain-specific sub-problems; ii) Interact: The agent then interacts with domain-specific expert models by repeating the ''ask-answer'' process to progressively obtain different domain-specific knowledge; iii) Integrate: The agent finally integrates all the acquired domain-specific knowledge to accurately address the medical reasoning problem. We validate the effectiveness of our method on the task of difference visual question answering for X-ray images. The experiments demonstrate that our zero-shot prediction achieves state-of-the-art performance, and even outperforms the fully supervised methods. Besides, our approach can be incorporated into various LLMs and multimodal LLMs to significantly boost their performance.
We evaluated our framework on the public dataset MIMIC-Diff-VQA, please refer to their repo and Physionet for data access.
- Convert Image Run the following code to create the PNG images that we are gonna work with.
python data_preprocess/converter.py -p <input_path_to_mimic_cxr_jpg> -o <output_path_to_mimic_cxr_png>- Prepare Dataset Modify line 25-26 in preprocess_data.py in the data_preprocess file, and run the script to generate the training, validation and test data accordingly.
python data_preprocess/preprocess_data.pyIt will generate the data for each type of questions in src/data.
├── data
│ ├── data_abnormality
│ │ ├── cache
│ │ │ ├── train_target.pkl
│ │ │ ├── vak_target.pkl
│ │ │ ├── test_target.pkl
│ │ │ ├── trainval_ans2label.pkl
│ │ │ ├── trainval_label2ans.pkl
│ │ ├── glove
│ │ │ ├── glove.6B.300d.txt
│ │ ├── maml
│ │ │ ├── t0_pretrained_maml_pytorch_other_optimization_3shot_newmethod.pth
│ │ │ ├── t1_pretrained_maml_pytorch_other_optimization_3shot_newmethod.pth
│ │ │ ├── t2_pretrained_maml_pytorch_other_optimization_3shot_newmethod.pth
│ │ │ ├── t3_pretrained_maml_pytorch_other_optimization_3shot_newmethod.pth
│ │ │ ├── t4_pretrained_maml_pytorch_other_optimization_3shot_newmethod.pth
│ │ │ ├── t5_pretrained_maml_pytorch_other_optimization_3shot_newmethod.pth
│ │ ├── dictionary.pkl
│ │ ├── embed_tfidf_weights.pkl
│ │ ├── glove6b_init_300d.npy
│ │ ├── imgid2idx.json
│ │ ├── pretrained_ae.pth
│ │ ├── trainset.json
│ │ ├── valset.json
│ │ ├── testset.json
│ ├── data_view
│ ├── data_level
│ ├── data_type
│ ├── data_presence
│ ├── data_location
│ ├── data_all_and_diffFor each type of questions regarding abnormalities, level, location, type, view, presence, train a seperate domain expert with the following command. It will save the checkpoints in src/save_models, which will be utilized later.
python main.py --use_VQA --VQA_dir data_level --maml --autoencoder --feat_dim 64 --img_size 84 --maml_model_path pretrained_maml_pytorch_other_optimization_3shot_newmethod.pth --maml_nums 2,5 --model BAN --lr 0.01 --seed 2104 --output saved_models/MMQ_BAN_MEVF_levevl --load
We also include the detected abnormalities in save_models, which is the prediction provided by a 121-densenet adapted from CheXclusion.
Modify line 23 with your openai key and run the bash script if you use.
run_demo.sh
Or directly run the Python command
python demo_mentors_final.py --use_VQA --VQA_dir data_all_with_diff --maml --autoencoder --feat_dim 64 --img_size 84 --maml_model_path pretrained_maml_pytorch_other_optimization_3shot_newmethod.pth --input saved_models/MMQ_BAN_MEVF_all --maml_nums 2,5 --model BAN --epoch _best --batch_size 32 --load --split train