An implementation of the paper Interpretable Deep Learning based Hippocampal Sclerosis Classification.
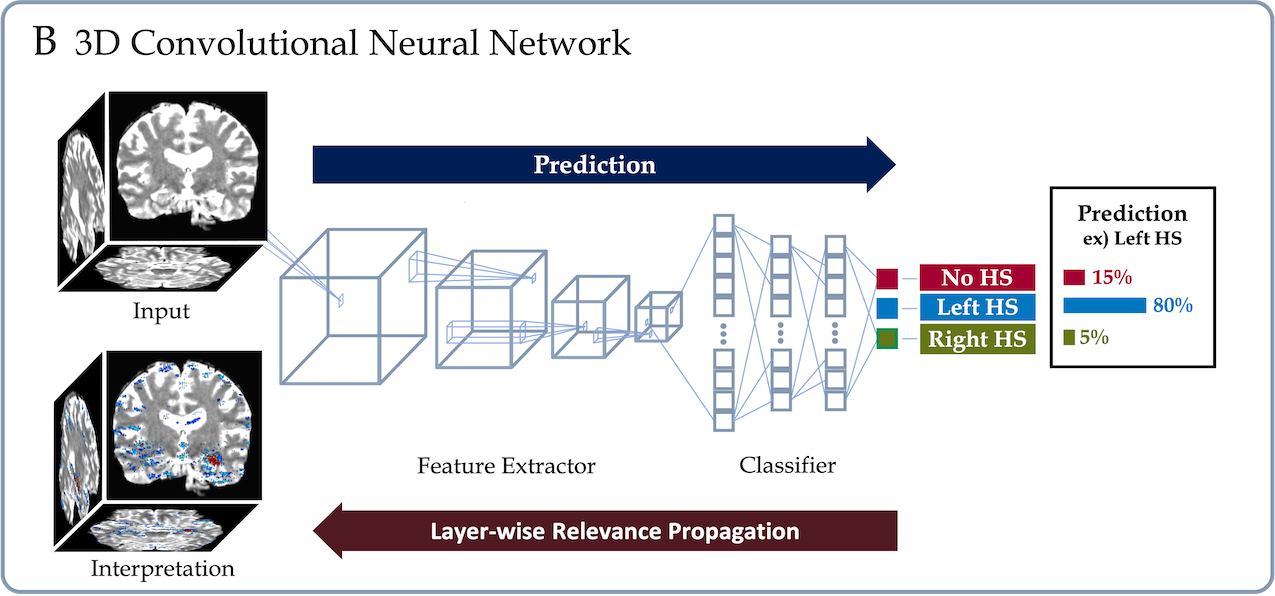
This repo contains Keras and PyTorch implementations of a deep learning framework that delineates interpretable Hippocampal Sclerosis(HS) prediction from Magnetic Resonance Imaging. We implemented a framework using a 3D convolutional neural network that was trained in an end to end fashion and generated an interpretation for the prediction through the Layerwise-relevance propagation method (Bach et al., 2015). See below.
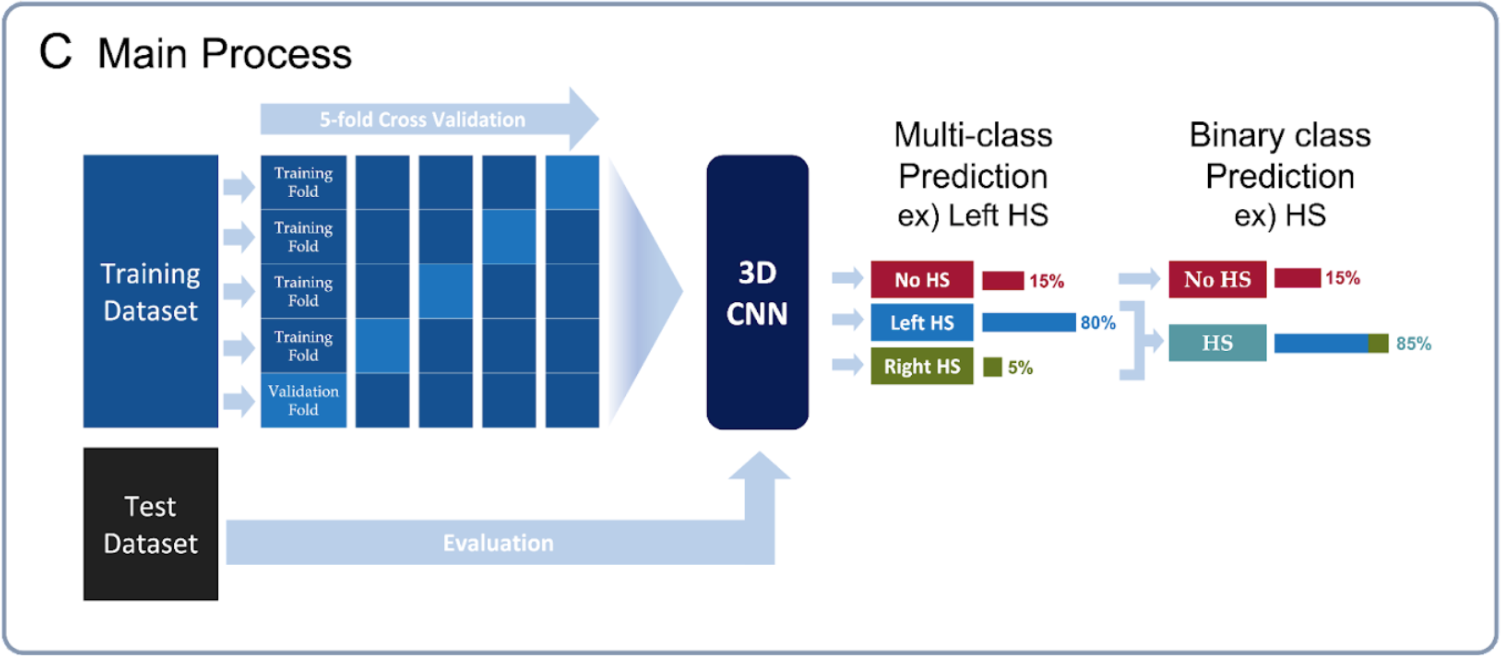
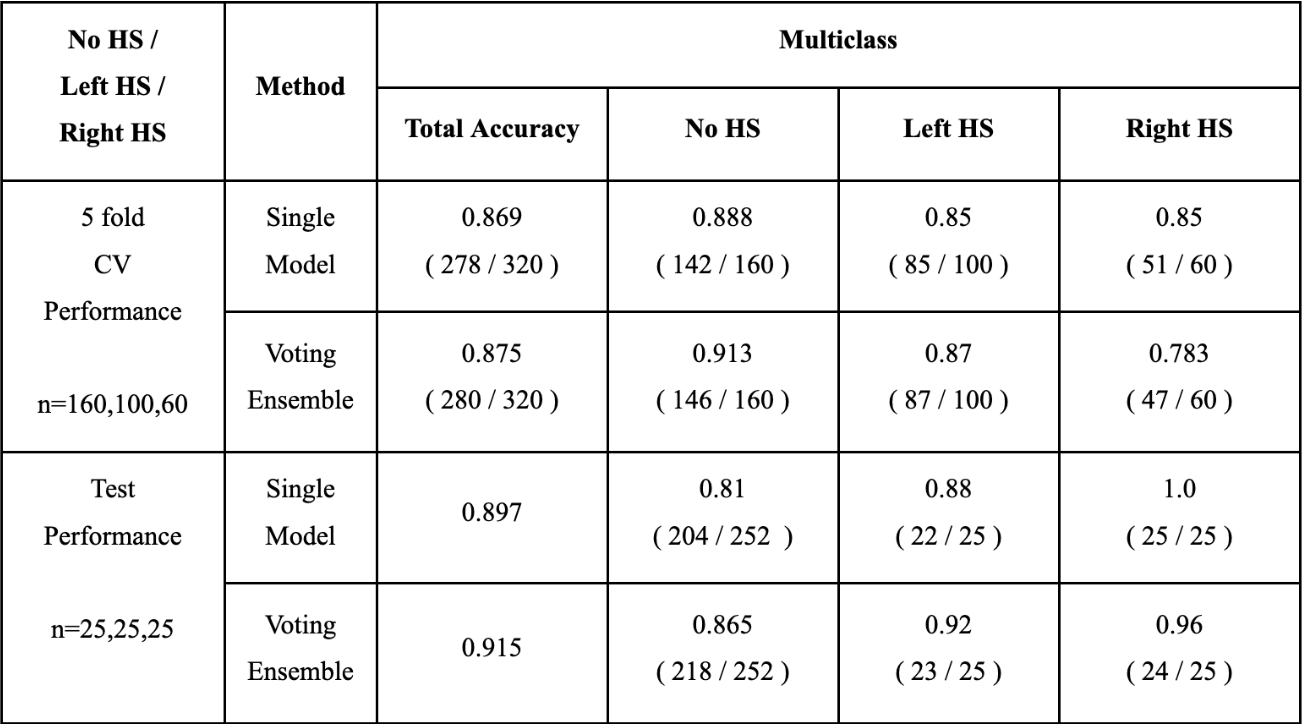
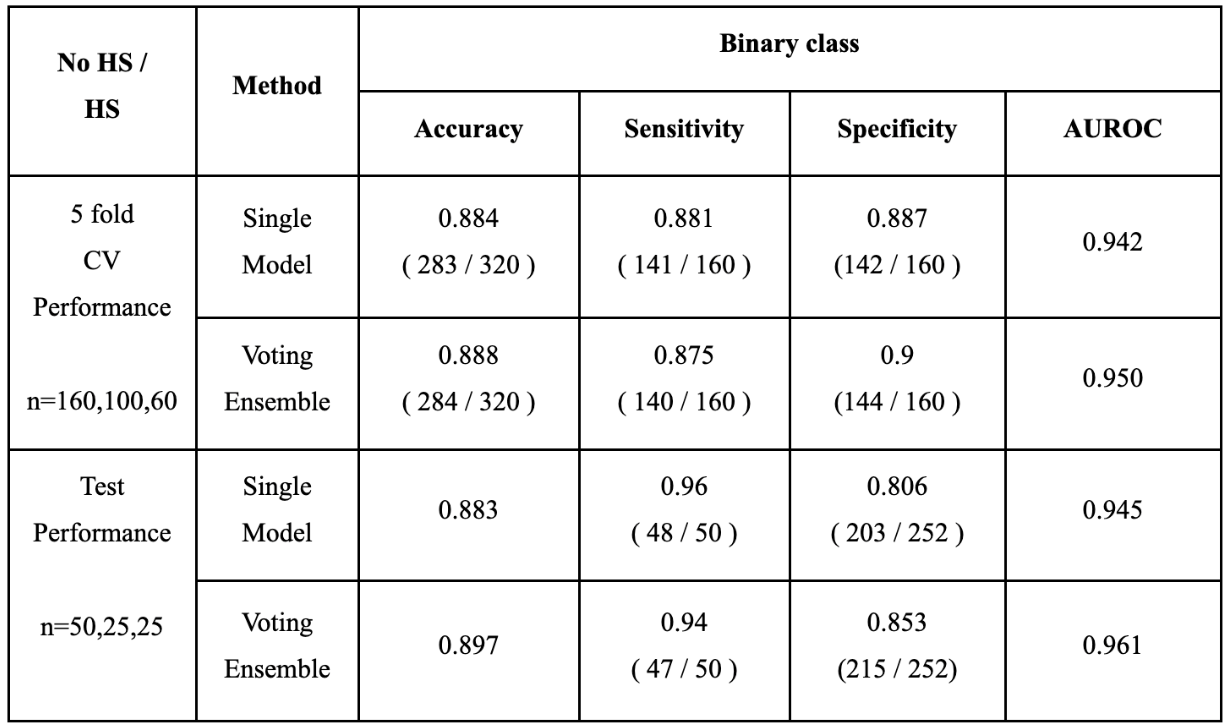
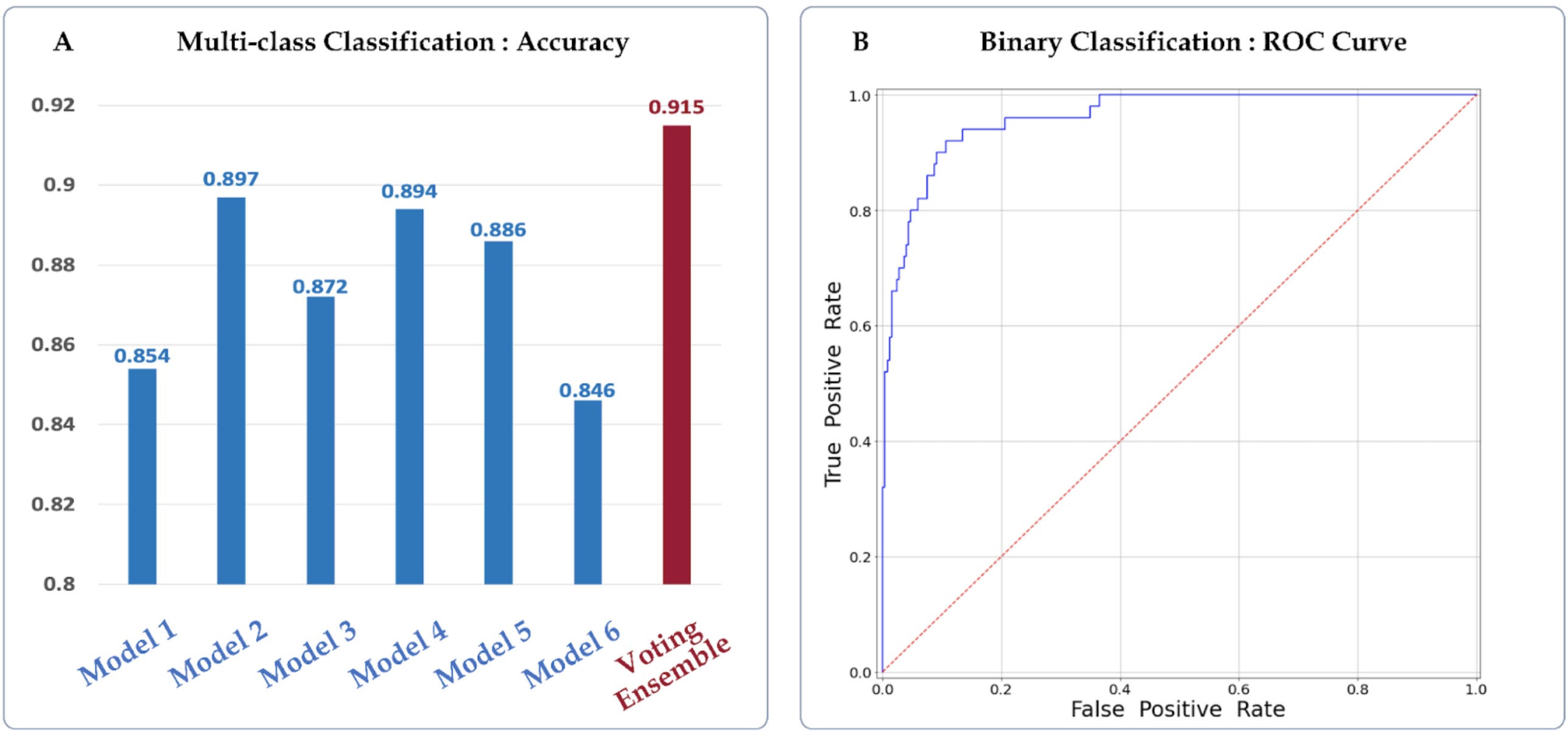
The CNN model was developed on the HS dataset that was provided by the Seoul National University Hospital and evaluated through both 5-fold cross validation (CV) and using a separate balanced test dataset. By combining the prediction probabilities for the Left and Right HS classes and comparing it with the probability for the No HS class, we not only considered a multi-class classification setting, but also an easier binary classification setting in which the goal is to simply determine whether a person has HS or not.
The training dataset was comprised 160 controls, 100 Left and 60 Right HS participants, and the test dataset was consisted of 252 control, 25 Left and 25 Right HS participants. Since the test dataset was imbalanced, we randomly sampled 25 and 50 participants from the control group 100 times to make a balanced test datasets for multi-class and binary classification. Since the test set was imbalanced, we took an average of the accuracy achieved within each group to measure a balanced accuracy for multiclass and binary classifications. We employed the voting ensemble to improve the prediction accuracy. For the test dataset, we plot the accuracies among several single models and their ensemble, and the ROC curve of the Voting Ensemble method for the multi-class and binary classification settings, respectively.
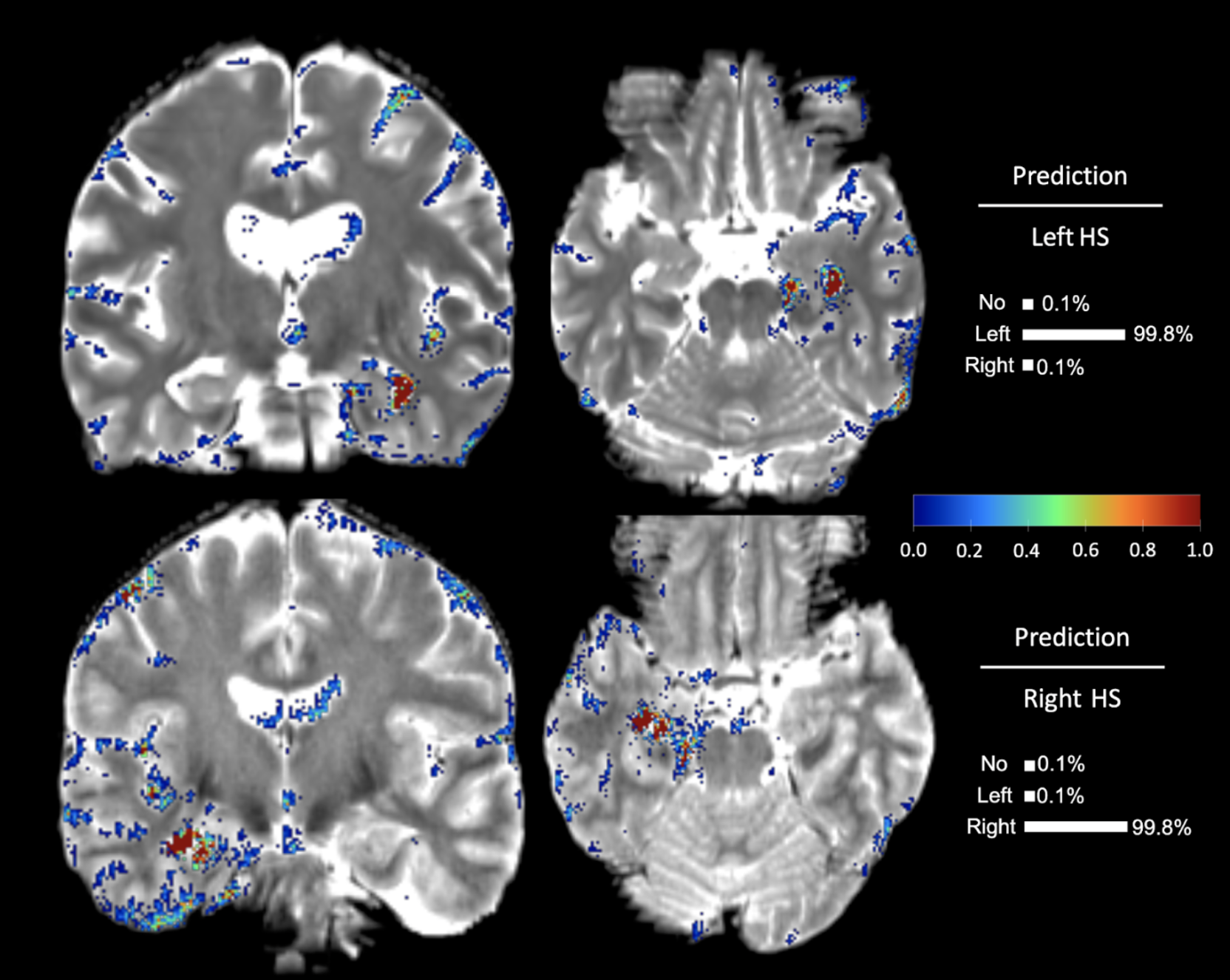
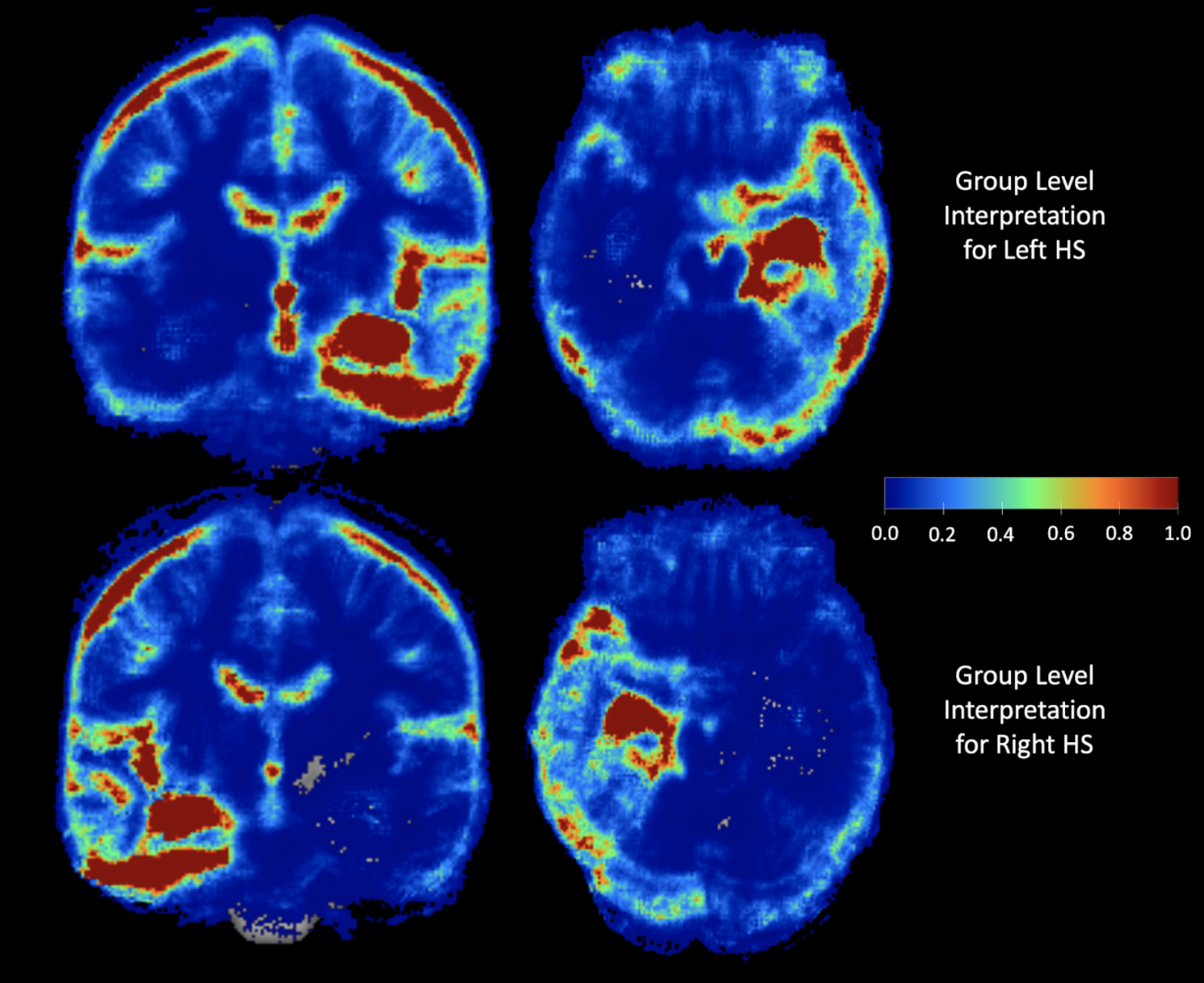
Once the model makes a prediction for a given individual 3D brain image, LRP recursively runs the relevance propagation step, which is similar to the standard back-propagation for neural networks training, to decompose and distribute the final prediction score to each input voxel. The decomposed score, dubbed as the relevance score, of LRP represents the importance of each voxel for the given prediction, and we obtain the saliency-map of the important voxels by visualizing the relevance scores higher than a threshold. We presented the interpretation both in individual and group-level.
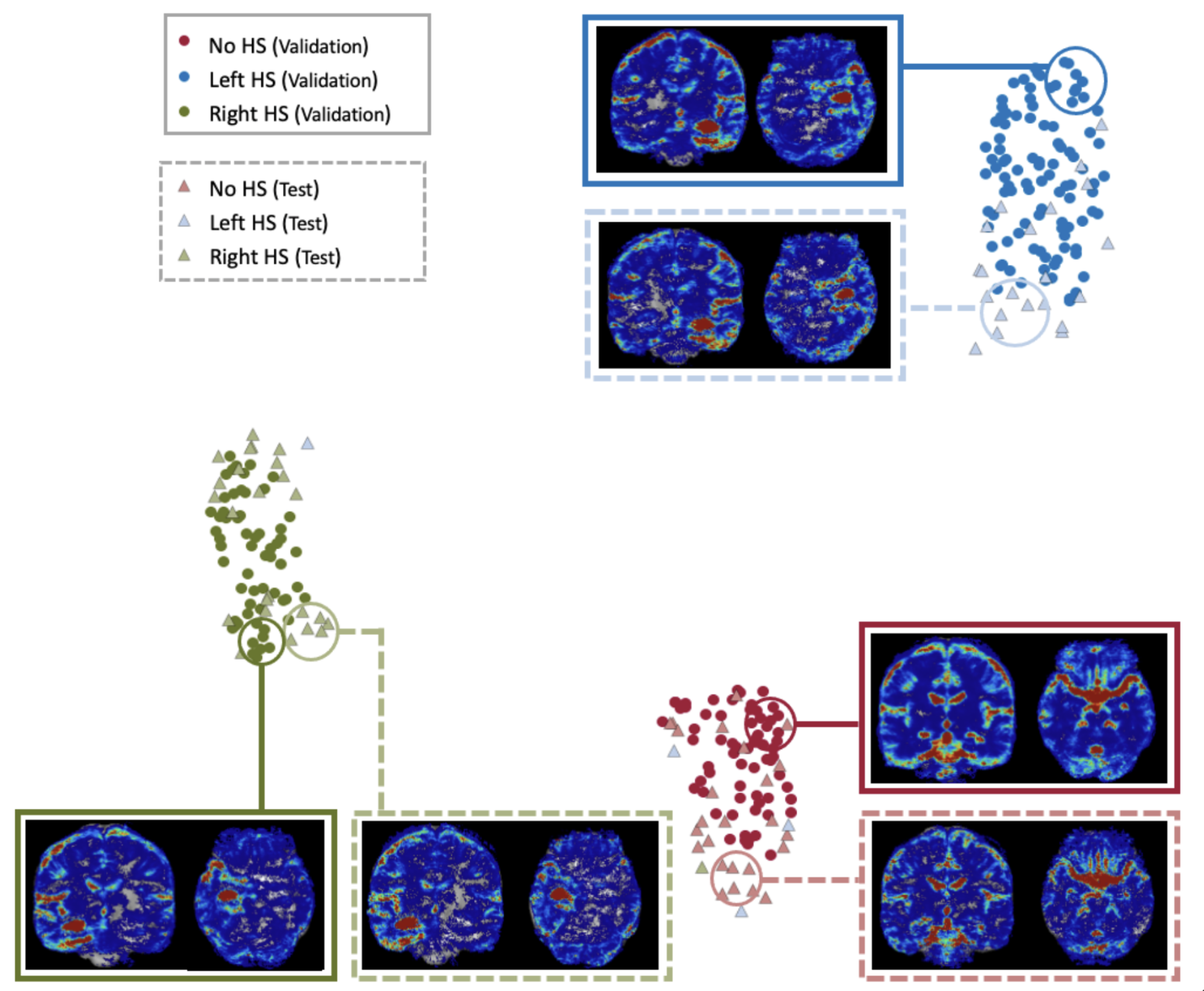
We applied a dimension reduction method called Uniform manifold approximation and projection (Mclnnes et al., 2018) to the output of the last convolutional layer in our 3D CNN. Figure 4 shows the projection of the feature embeddings to the two-dimensional space, and the solid circles and light triangles correspond to the training and test data samples, respectively.
Please refer to our paper for more details.
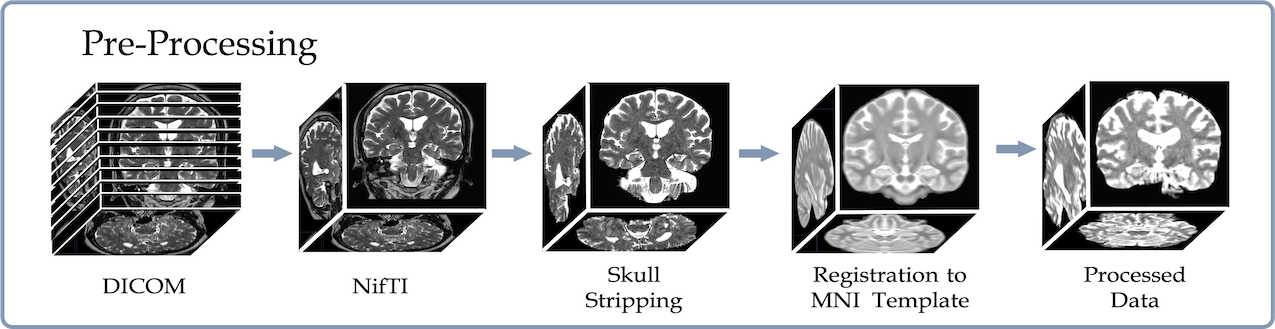
We trained, validated and tested the framework using the Hippocamapl Sclerosis (HS) dataset from the Seoul National University Hospital. To download the raw data, please contact those affiliations directly. We provided all data preprocessing manuscripts in "./Data_preprocessing/" folder.
We used the MRIcroGL program to convert the MRI file in Dicom format to Nifti format
-
step1: Deoblique the data using AFNI 3drefit function (need AFNI to be installed).
Fix up errors made when using to3d.
3drefit -deoblique $entry -
step2: Reorient the data using AFNI 3dresample function (need AFNI to be installed).
Reorient the direction as same as the standard brain template to use.
3dresample -orient LPI -prefix $LPI_image -inset $entry -
step3: Bias field correction using AFNI 3dUnifize function (need AFNI to be installed).
Remove the shading artifacts Approximately uniformized the white matter (WM) intensity across space and scaled to peak at about 1000.
3dUnifize -input $LPI_image -prefix $BFcor_image -T2 -clfrac 0.3 -T2up 99.5 -
step4: Skull stripping using AFNI 3dSkullStrip and 3dcalc function (need AFNI to be installed).
Generate a skull stripped mask and apply element-wise multiplication with the original input. Preserve not only the range of values of the brain but also some low-intensity voxels.
3dSkullStrip -mask_vol -input $BFcor_image -prefix $mask_image) 3dcalc -a $BFcor_image -b $mask_image -expr 'a*step(b)' -prefix $SS_imageWe provided this bash pipeline (Data_preprocessing/Preprocessing.sh) to perform this step :
bash Preprocessing.sh
3. Registration onto the standard brain template using flirt function (need FSL to be installed):
Register the skull stripped brain onto the the MNI brain template. Use flirt function (need FSL to be installed.)
flirt -in $SS_image -ref zMNI_1x1x1_brain.nii.gz -out $image_MNI -omat $MNI_mat -dof 12
We provided this bash pipeline (Data_preprocessing/Registration_MNI.sh) to perform this step :
bash Registration_MNI.sh
Background outside the brain still exists in the MRI. The redundancy increases the data volume and correspondingly makes the model consider unnecessarily additional region. We explored the data to see the general boundary of the background region in all 3 dimensions and cut the data in the extent. To do this step, run a python file in "/Data_preprocessing" folder :
python back_remove.py
The tool was developped based on the following packages:
- NumPy (1.15.4 or greater).
- Tensorflow (1.12.0).
- Keras (2.2.0).
- PyTorch (1.4.0 or greater).
- Nibabel (3.0.2)
- AFNI
- FSL
Please note that the dependencies may require Python 3.5 or greater. It is recommemded to install and maintain all packages by using conda or pip. For the installation of GPU accelerated PyTorch, additional effort may be required. Please check the official websites of PyTorch and CUDA for detailed instructions. The keras code was implemented on this Docker.
For Keras,
python HS_main.py --CONTROLTYPE CLRM --SETT SIG --AUG hflip --KERNEL_SEED 1 --TRIAL 1 --DATATYPE 64 --BATCH 16 --MODEL 3D_BASIC5124 --FC1 64 --FC2 64
For Pytorch,
python main.py --CONTROLTYPE CLRM --SETT SIG --AUG hflip --KERNEL_SEED 3 --TRIAL 03 --DATATYPE 60 --BATCH 42 --MODEL 3D_5124 --step 50 --TALK M5124_3 --drop 1
Descriptions for each option are described in the "/Args/argument.py".
Model weights will be saved in "/saveModel" folder and the evaluation result will be saved in "/log" and "/graph" folder.
For Keras,
python HS_ensemble.py --AUG hflip --TRIAL 9 --SETT SIG --DATATYPE 64 --CONTROLTYPE CLRM --EnsMODE AVR --KERNEL_SEED 1
For Pytorch,
python ensemble.py --CONTROLTYPE CLRM --SETT SIG --AUG hflip --KERNEL_SEED 333 --TRIAL 45 --DATATYPE 60 --BATCH 42 --MODEL 3D_5124 --step 50 --TALK AVR_ensemble --K1 2 --K2 14 --K3 19 --K4 23 --K5 33 --EnsMODE AVR
EnsMODE controls the ensemble mode.
Two options are given : 'AVR' for average ensemble and 'VOT' for voting ensemble
For Keras,
python HS_LRP.py --SETT SIG --TRIAL 1 --RULE lrp.z --KERNEL_SEED 1 --CONTROLTYPE CLRM --AUG hflip --DATATYPE 64 --PERCENT 5 --LRPMSG Directory_name
For Pytorch,
python lrp.py --SETT SIG --TRIAL 03 --RULE lrp.z --KERNEL_SEED 14 --CONTROLTYPE CLRM --AUG hflip --DATATYPE 60 --PERCENT 0 --LRPMSG Directory_name --MODEL 3D_5124
Controls the cutoff values using the PERCENT.
In both codes, we set all voxels with negative relevances with zero and normalize them to be in range 0 ~ 100.
In "keras/graph" folder,
python dim_reduction.py --SEED 3 --INTEREST 16 --PERP 85 --EPSI 200 --STEP 3000 --SELECT UMAP --TRIAL 13 --CONTROLTYPE CLRM --SETT SIG
You may adjust the hyperparameters like perplexity or step to achieve different outcomes.
The result would be saved in "/keras/graph/UMAP" folder.