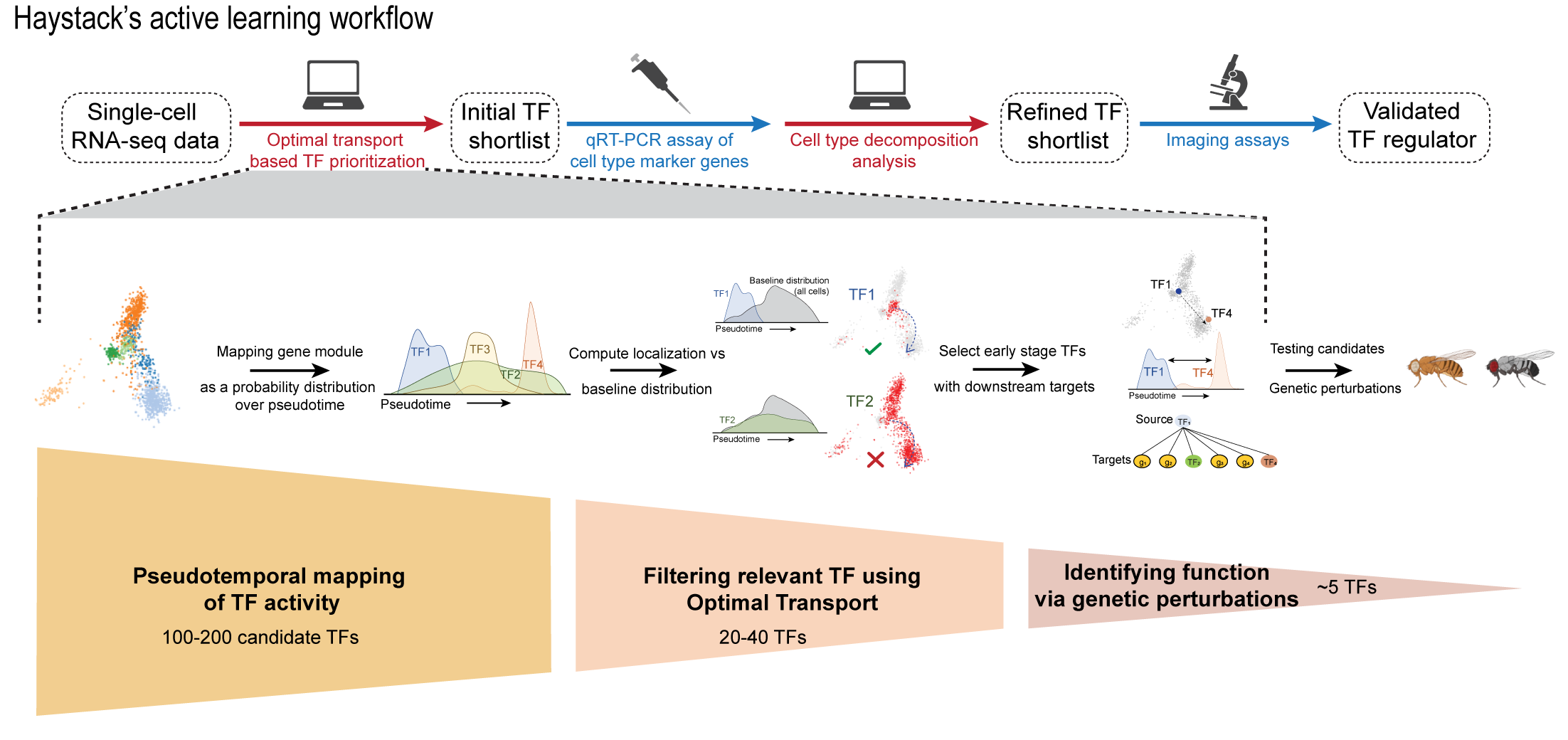
The explosive growth of single-cell transcriptomics (scRNA-seq) has made it convenient to study differentiation trajectories and generate hypotheses about transcription factors that may be playing a causal role in differentiation. However, the bottleneck has now shifted to the validation of these hypotheses. Since most scRNA-seq studies are observational, with lineage-determining TFs predicted by correlational analyses, perturbational experiments are required to test these predictions and confirm causality. Yet such validations remain slow and expensive and are far outpaced by high-throughput scRNA-seq assays. Haystack is designed to optimally allocate limited experimental resources toward identify high-confidence scRNA-seq hypotheses and efficiently guide in vivo validations.
Haystack is described in the bioRxiv preprint, Optimal transport analysis of single-cell transcriptomics directs hypotheses prioritization and validation by Rohit Singh*, Joshua Li*, Sudhir Tattikota*, Yifang Liu, Jun Xu, Yanhui Hu, Norbert Perrimon, and Bonnie Berger.
Coming soon...