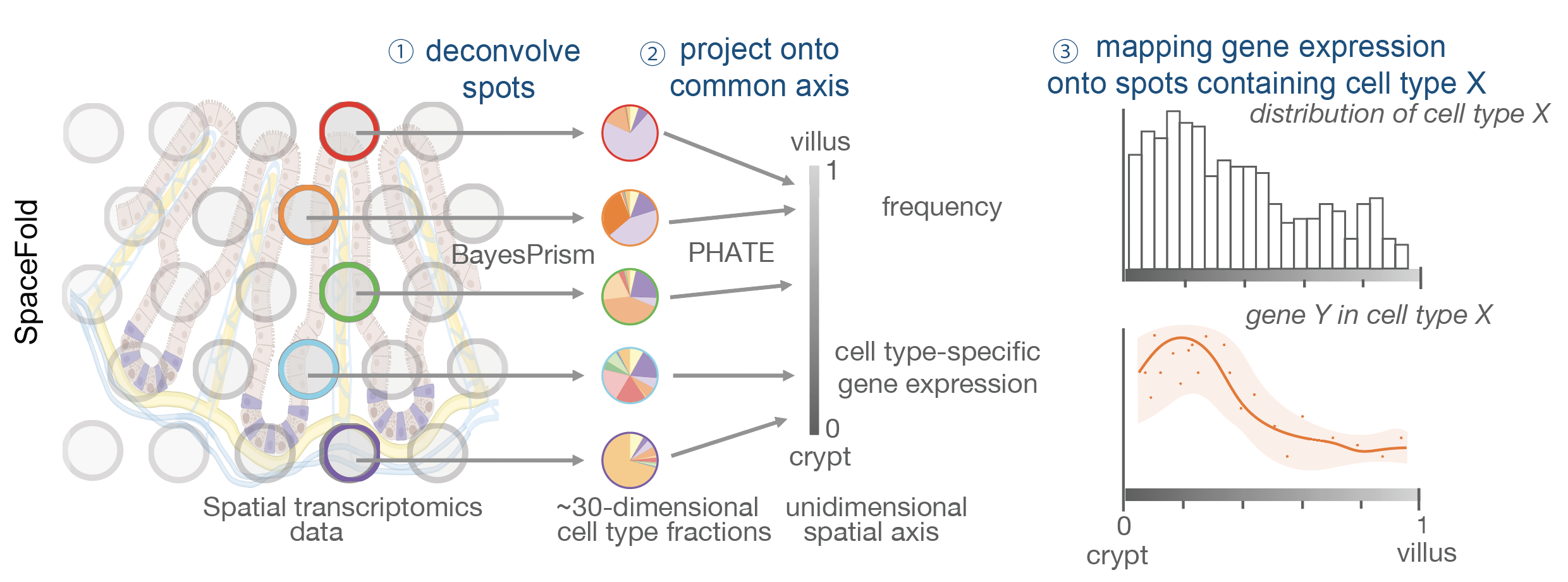
Mapping gene expression cartography from spatial transcriptomics for tissues with stereotypical structures.
What's new?
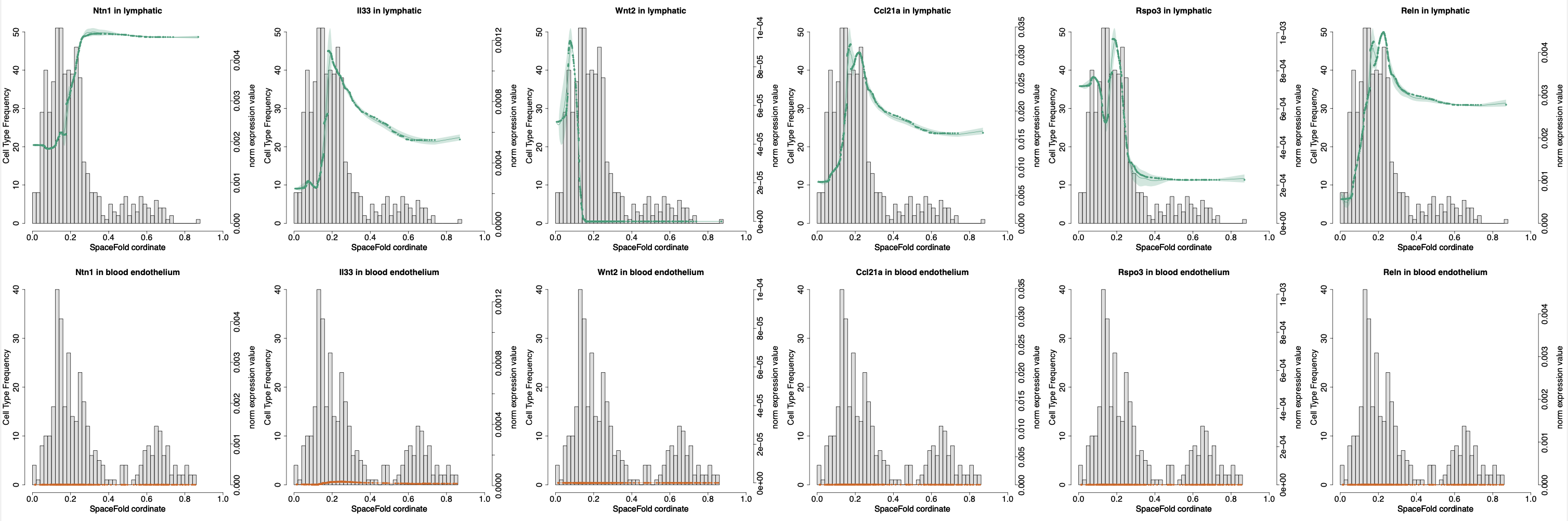
We developed a new doising method using data diffusion over the imputed SpaceFold axis to oversome sparsity of Visium data and further improve the resolution of cartography over the sliding window mean used by the v1.0 (the SpaceFold paper).
The new version v2.0 is compatible with the new BayesPrism package (https://github.com/Danko-Lab/BayesPrism).
A comparison between v1.0 and v2.0:
Space Ranger output and the h5ad files of the scRNA-seq data can be downloaded from https://doi.org/10.6084/m9.figshare.19715029 .
We provide a computational gateway to run SpaceFold on a HPC server, and visualize SpaceFold cartography for mouse small and large intestine tissues. Additionally, this gateway portal also provides data explorer for the scRNA-seq data from mouse small and large intestine tissues.
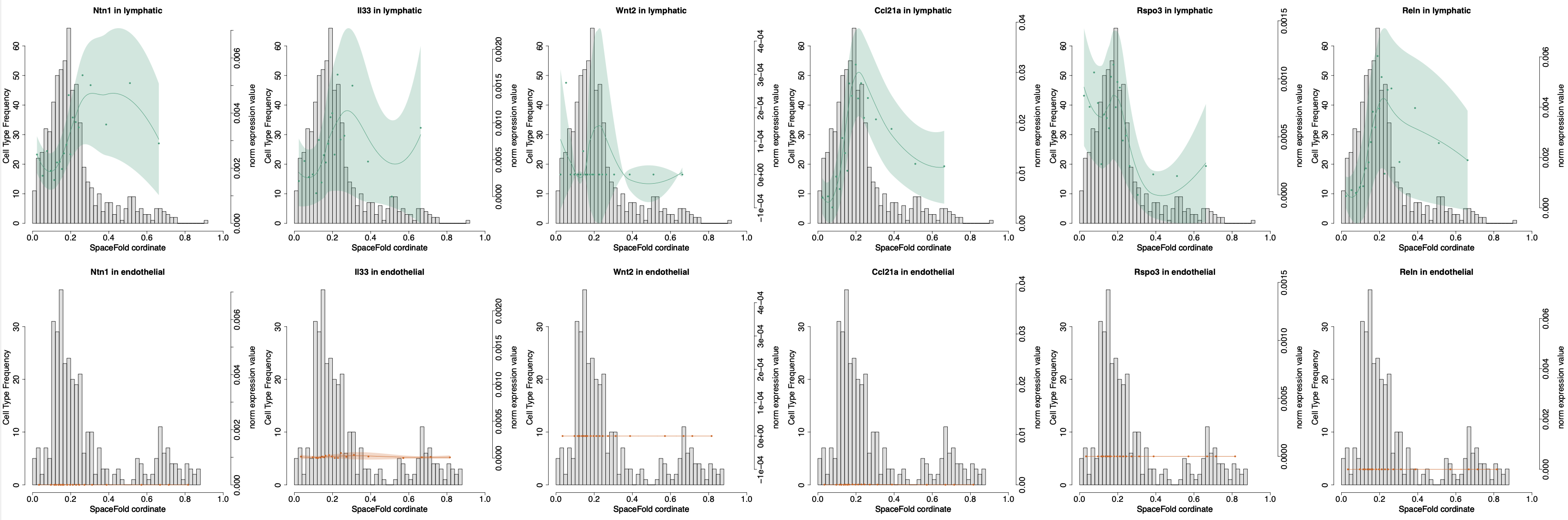
Lymphatics act as a signaling hub to regulate intestinal stem cell activity
https://doi.org/10.1016/j.stem.2022.05.007
-
R packages:
BayesPrism, expm, msir, mixtools, mclust, phateR, RColorBrewer, beeswarm, dplyr
-
BayesPrism can be installed by:
library("devtools");
install_github("Danko-Lab/BayesPrism/BayesPrism")
-
Recommended: pdfjam: https://github.com/rrthomas/pdfjam
Each panel will conatin the expression cartography of one gene in one cell type along the SpaceFold axis. If pdfjam is installed, output will be a pdf file with all panels concatenated as a 2D matrix. Otherwise, output will be a pdf file with one panel on each page.
-
User needs to make sure that the python end of PHATE is installed properly: https://github.com/KrishnaswamyLab/phateR . Also check the python version with PHATE installed is linked by reticulate properly (use use_python and use_condaenv to configure). Otherwise run.phate will report error.
-
If all dependent packages and commands have been installed, please use the following codes to install/update the package in R terminal.
library("devtools");
install_github("dpeerlab/SpaceFold/SpaceFold")
library(SpaceFold)
See the vignette.R for details.
- What are the assumptions made by SpaceFold?
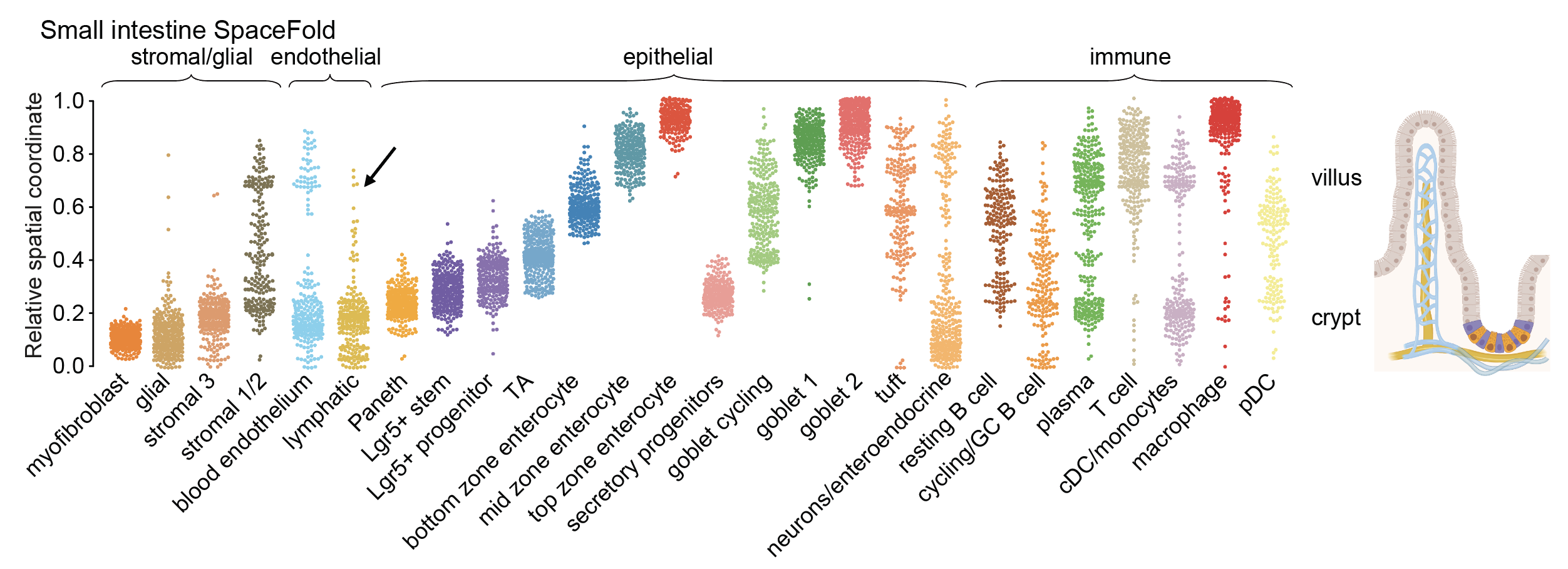
SpaceFold assumes that 1) the tissue has an underlying 1D structure; 2) it is a stereotypical structure that is repeatedly sampled by ST; 3) the cell type fraction of a spatial spot is sufficient in inferring its physical cordinate along the 1D axis.
- R vignette:
- vignettes.html
To view, please git clone the repository and open the html files using your browser.
- R manual: (Coming soon)