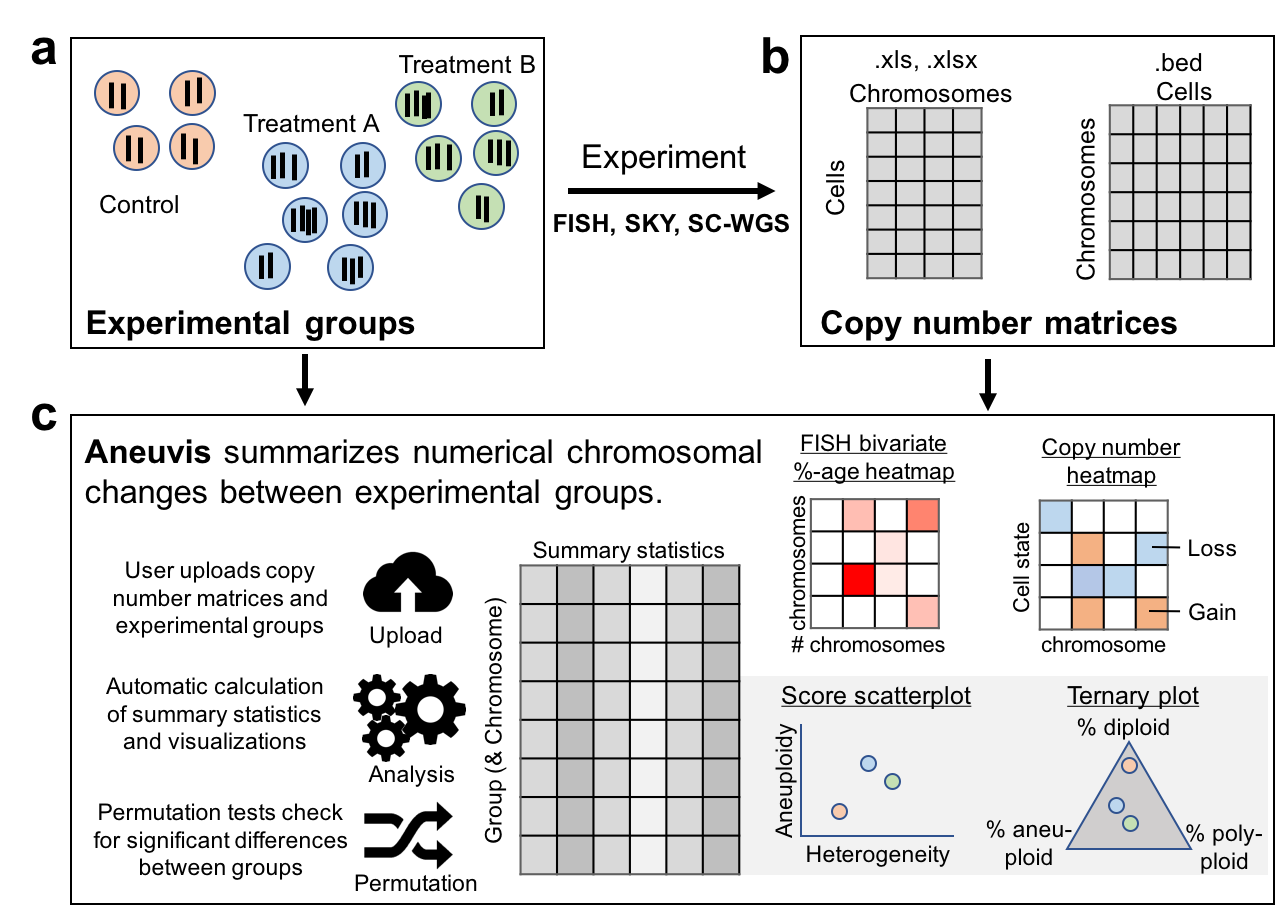
aneuvis is a web-based tool to automatically analyze numerical chromosomal variation in single cells.
A video tutorial demonstrating the usage of aneuvis is available.
A schematic of the features of aneuvis is shown below.
aneuvis is available to use at dpique.shinyapps.io/aneuvis/
To run aneuvis locally, first clone this repo to your computer:
git clone https://github.com/dpique/aneuvis.git
Then, change directories into the aneuvis directory:
cd aneuvis/
Lastly, load the app. Ensure that Shiny and the required libraries are installed (see header of app.R for required R packages.)
R -e "shiny::runApp('app.R')"
aneuvis was built using R version 3.4.3 and shiny version 1.0.5 and is freely available under a GPLv3 license.