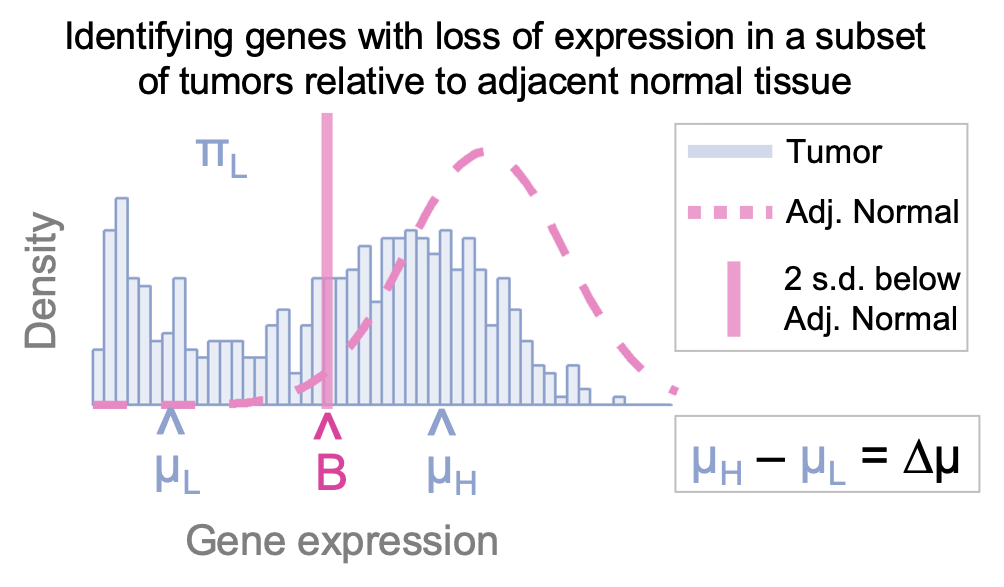
This package contains functions to detect nuclear hormone receptor (NHR) expression loss (receptLoss) in subsets of tumors relative to normal tissue.
-
receptLoss: the main function in this package.-
Input: 2 matrices of expression data, one from tumor and one from adj normal tissue.
-
Output: Table of summary statistics, with each row as a gene and each column a separate statistic. The table's rows are sorted by delta mu value, which reflects the degree of separation between tumor subgroups.
-
-
plotReceptLoss: A function to visualize the output from receptLoss for a particular gene.
It also includes a dataset consisting of all known NHRs:
receptLoss is complementary to oncomix. Whereas oncomix detects the gain of gene expression in subsets of tumors relative to normal tissue, receptLoss detects the loss of gene expression in subsets of tumors relative to normal tissue.
## Install the development version of receptLoss from github
devtools::install_github("dpique/receptLoss",
build_opts=c("--no-resave-data", "--no-manual"),
build_vignettes=TRUE)
## open the vignette
vignette("receptLoss")