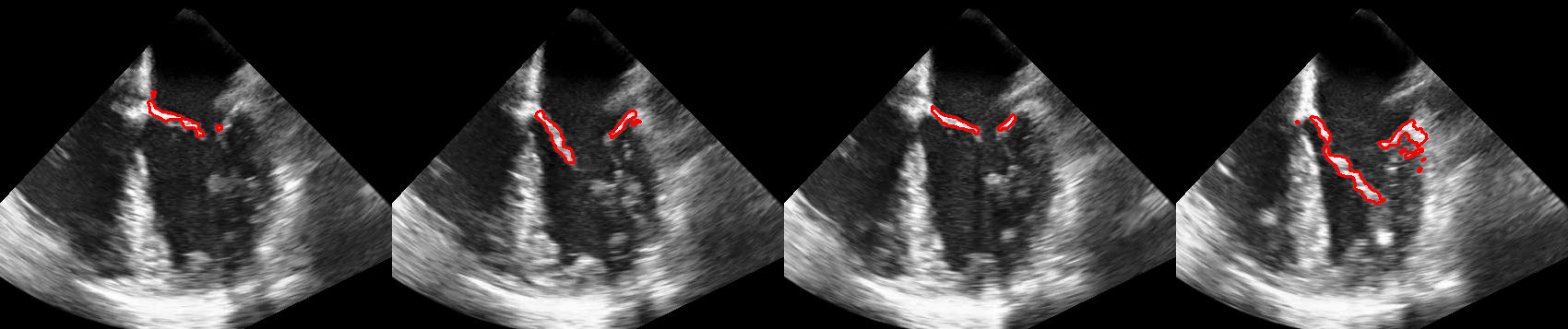
This repository is the official MATLAB implementation of Mitral Valve Segmentation using Robust Nonnegative Matrix Factorization.
Some results of the proposed methods are shown in: https://drive.google.com/drive/folders/1Scpy54x0-_zXpP3bXZIYJPitN2kMUyB1?usp=sharing
- Please insert the videos that should be used for mitral valve segmentation into the folder:
./data/original/ - Create the ground truth data by running the file
./tools/createGroundTruth.m, which will be saved into./data/ground_truth/and./data/ground_truth_window/ - Run
./main.mto start the algorithm - Run
./tools/showResults.mto analyze the segmentation results
- To run one of the Robust Nonnegative Matrix Factorization (RNMF) methods, please run
ids = rnmf([methodname], [numberofvideos]); - Instead of methodname, insert one of the following methods: 'robustNMF', 'robustBreg', 'robustNMF_excludeWHS' or 'robustNMF_excludeWHS_Breg'
- Replace numberofvideos by the number of videos you want to segment
- Please adapt the settings for RNMF in
./rnmf.m
- To apply the segmentation methods, please run
segment('segmentCVPlus',[rnmfmethodname],ids,'postProcessing',[postprocessingsetting],'cropped', [windowingsetting]); - Instead of rnmfmethodname, insert the method of the following methods, on which reults you want to run the segmentation on: 'robustNMF', 'robustBreg', 'robustNMF_excludeWHS' or 'robustNMF_excludeWHS_Breg'
- Replace postprocessingsetting by: true, false or both, depending on if you would like to run the Refinement Step or not (or get results for both cases)
- Replace windowingsetting by true or false, depending on if you would like to apply the windowing approach of the window calculated in the RNMF step
- Please adapt the settings for the segmentation in
./segment.m