Orenogb
Visualization command for genomic data
Description
- just type simple command on your terminal
- beautiful visualization by ggbio and ggplot2
- semantic zoom
- eaily loading of transcriptome annotation
- search by gene Symbol
- choose species
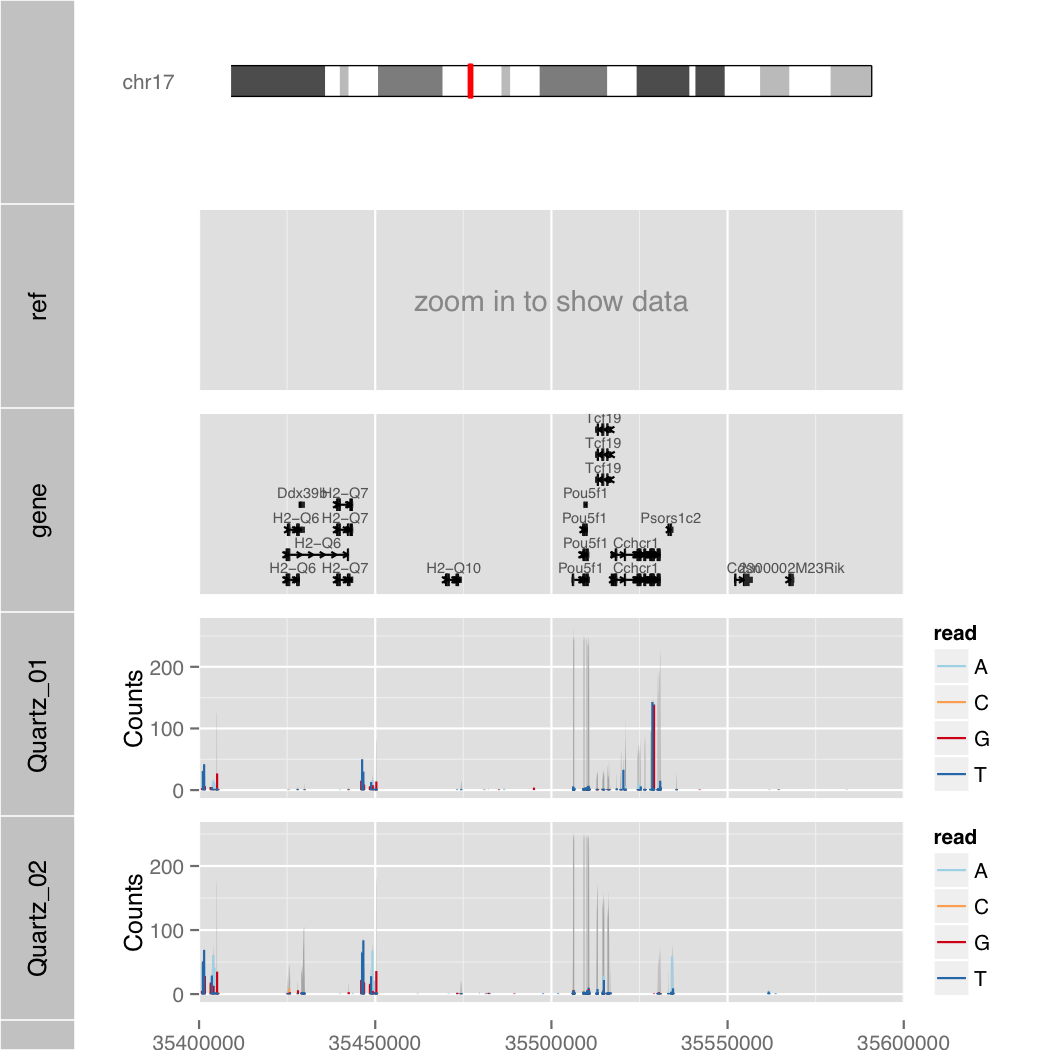
Demo
$ R --slave --vanilla -f orenogb.R --args coordination mm10 chr17 35400000 35600000 1 Quartz_01.bam,Quartz_02.bam demo/demo.pdf
Exponential notation
$ R --slave --vanilla -f orenogb.R --args coordination mm10 chr17 3.55e7+2880 3.55e7+16079 1 Quartz_01.bam,Quartz_02.bam demo/demo2.pdf
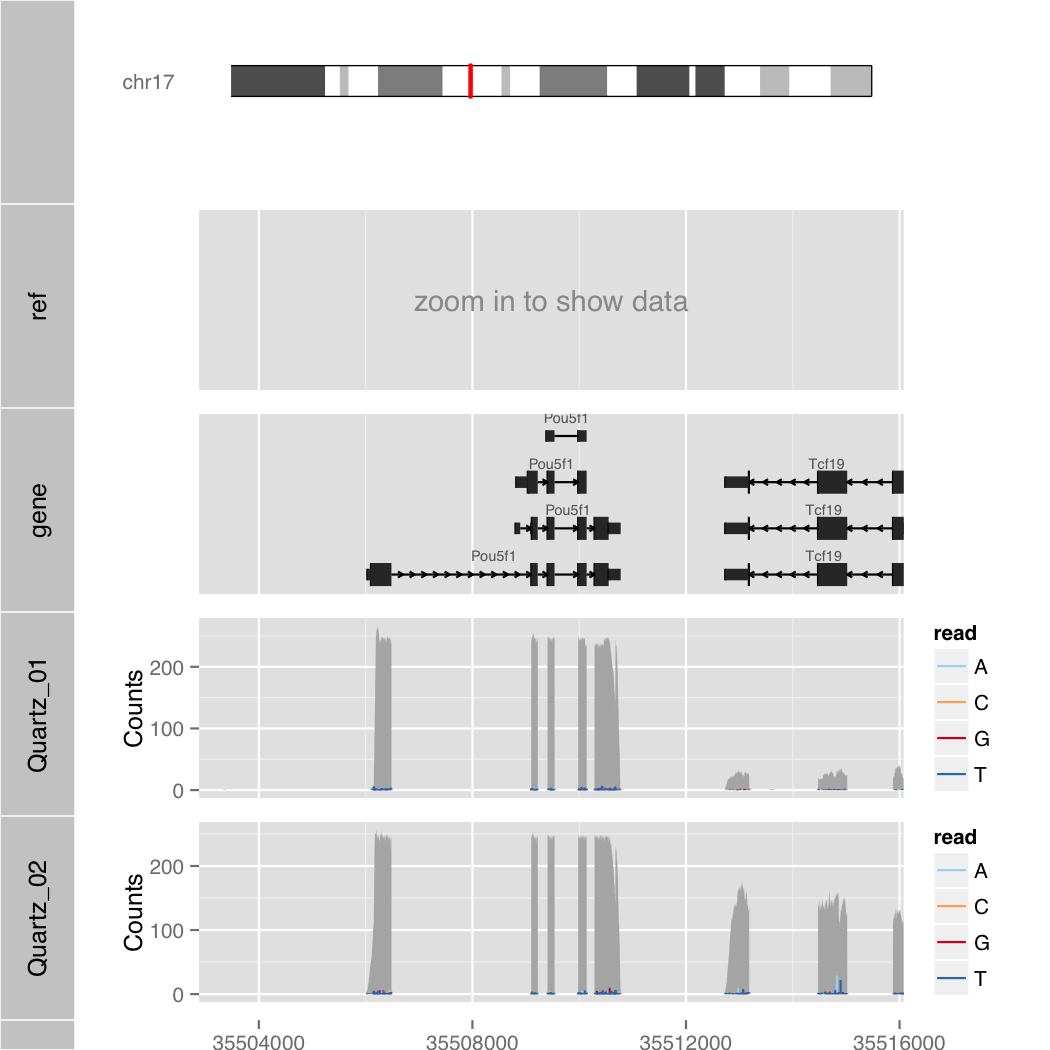
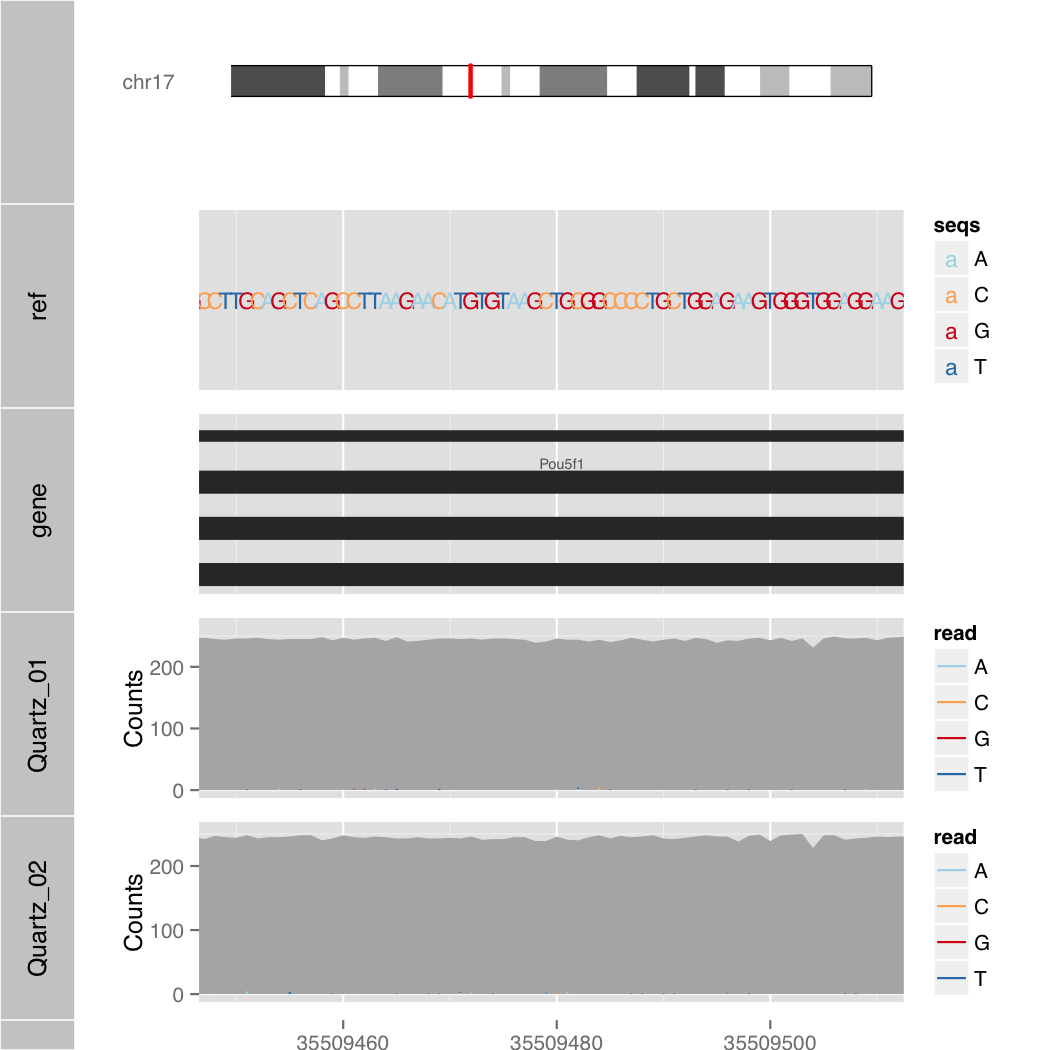
Semantic Zoom
$ R --slave --vanilla -f orenogb.R --args coordination mm10 chr17 35502880 35516079 1/200 Quartz_01.bam,Quartz_02.bam demo/demo3.pdf
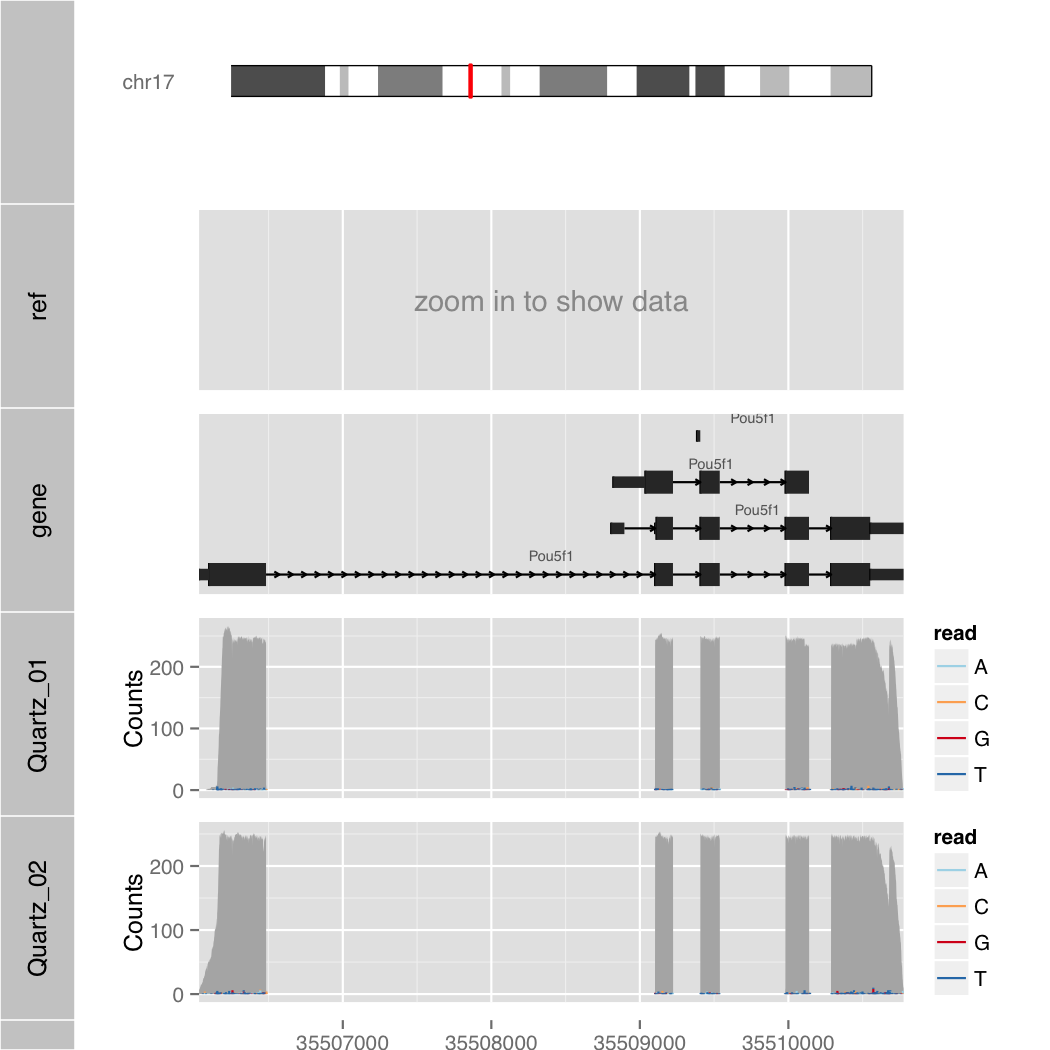
Search by Gene Symbol
$ R --slave --vanilla -f orenogb.R --args gene mm10 Pou5f1 1 Quartz_01.bam,Quartz_02.bam demo/demo4.pdf
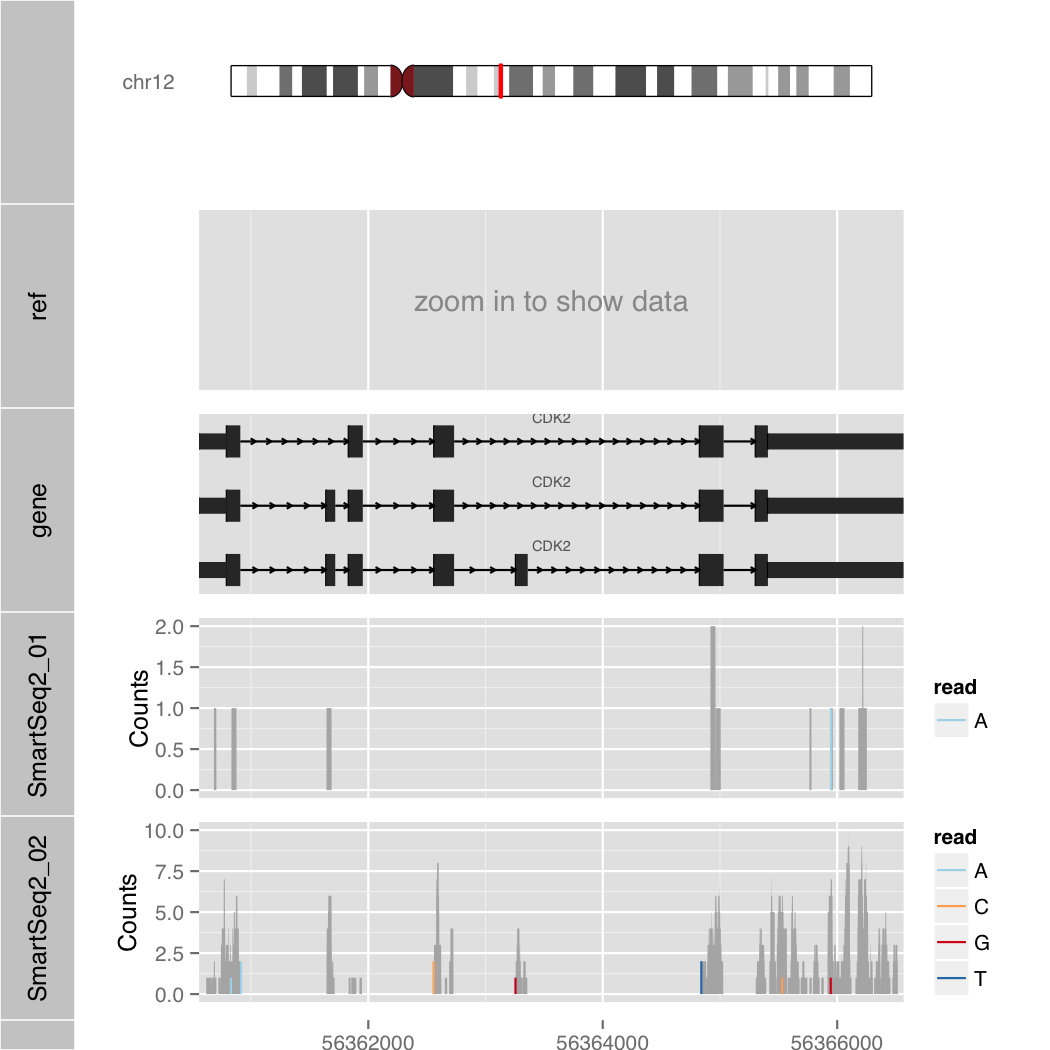
Human Genome
$ R --slave --vanilla -f orenogb.R --args gene hg19 POU5F1 1 Smart-Seq2_01.bam,Smart-Seq2_02.bam demo/demo5.pdf
Requirement
- R
- Bioconductor Software Packages
- ggbio
- GenomicRanges
- GenomicAlignments
- Bioconductor Annotation Packages
- Mus.musculus, Homo.sapiens, ...
- BSgenome.Mmusculus.UCSC.mm10, BSgenome.Hsapiens.UCSC.hg19, ...
Usage
$ R --slave --vanilla -f orenogb.R --args [mode] [genome] [chr] [start bp] [end bp] [zoom] [bam1,bam2,...] [output file]
Install
$ git clone git@github.com:dritoshi/orenogb.git
$ cd orenogb
$ sudo R
R> source("http://bioconductor.org/biocLite.R")
R> biocLite(c("ggbio", "GenomicRanges", "GenomicAlignments")
R> biocLite(c("Mus.musculus", "BSgenome.Mmusculus.UCSC.mm10"))
R> biocLite(c("Homo.sapiens", "BSgenome.Hsapiens.UCSC.hg19"))
ToDo
- choose species
- wapper by shell script
- unit test
- deamonaize R script
- draw sample name