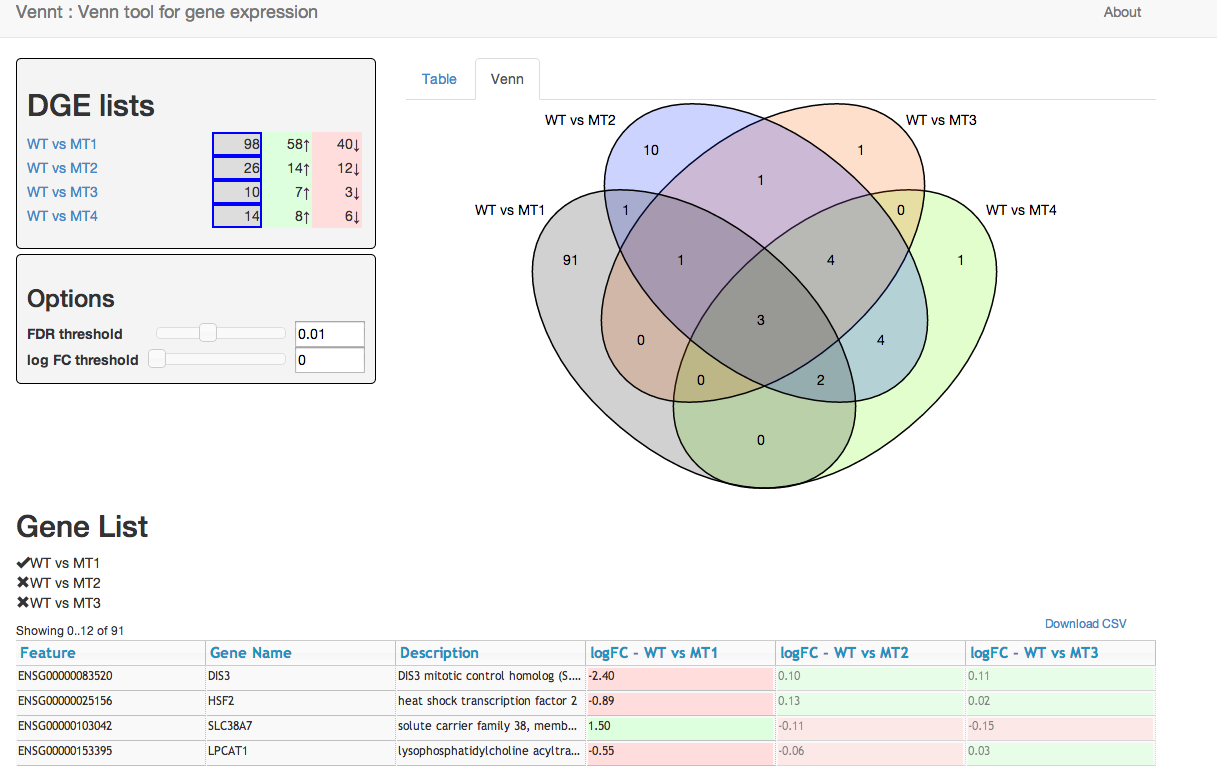
- Dynamic Venn diagrams for exploring lists of differential expressed genes
Try a Live Demo
Download this python script vennt.py (requires python >=2.7). Then simply run:
python vennt.py --cuffdiff gene_exp.diff > my-vennt.html
This will create a single html file that can be shared.
Generate a single CSV file with all your gene lists. Each row of the CSV should contain information about a gene including the log fold change, and the adjust p-value. Use a single column to specify the gene-list (see the example below). Each gene must have a unique identifier, which is used to find the corresponding genes in the different gene lists.
Download this python script vennt.py (requires python >=2.7). Then, if your CSV column names match the defaults, simply run it as follows:
python vennt.py gene-lists.csv > my-vennt.html
You may specify alternative column names, see python vennt.py -h for help. And read the settings list below for more information on the columns.
Creating a single HTML file with all your gene lists embedded may be a problem due to the size of the resulting HTML file. In that situation, you can serve the gene-list CSV file from a web-server. Firstly, create your CSV as described above.
Download this html file. Then, put it and your CSV file on a web-server. (For local testing you can use python -mSimpleHTTPServer.)
You may need to specify some configuration if the defaults do not suffice, for example column names. These are configured in the html file.
Set these in window.venn_settings in your html file (or using the options to vennt.py).
csv_file- (default 'data.csv') Name of the CSV file containing the data to load. Must be on the same origin as the html file due to javascript's Same-origin policycsv_data- (default 'null') - This can be used to directly embed a CSV file rather than requesting via ajax. Note, setting this parameter will cause anycsv_fileto be ignoredkey_column- (default 'key') Name of the column specifying the gene-list.id_column- (default 'Feature') Name of the column specifying a unique identifier for the gene. This must be unique within each gene-list, because it is used to match up the genes between the different gene-lists.fdr_column- (default 'adj.P.Val') - Name of the column containing the adjusted p-value. (This is often a false-discovery rate.)logFC_column- (default 'logFC') - Name of the column containing the log-fold-change for each gene-list.info_columns- (default '[Feature]') - An array of column names to display to the user. This should contain useful information you want the user to see - such as a gene-id, perhaps common gene-name, or possibly a brief description.show_tour- (default true) - Show the Venn tour on page load (if not shown before)
For example, consider this is your csv file, which is called data.csv:
gene-list,id,Description,Gene Name,logFC,adj.P.Val
WT vs MT1,ENSG00000083520,DIS3 mitotic control homolog (S. cerevisiae),DIS3,-2.4,4.8e-10
WT vs MT1,ENSG00000025156,heat shock transcription factor 2,HSF2,-0.89,6.4e-05
WT vs MT1,ENSG00000103042,"solute carrier family 38, member 7",SLC38A7,1.5,6.4e-05
WT vs MT2,ENSG00000083520,DIS3 mitotic control homolog (S. cerevisiae),DIS3,-2.4,4.8e-10
WT vs MT2,ENSG00000025156,heat shock transcription factor 2,HSF2,-0.89,6.4e-05
WT vs MT2,ENSG00000103042,"solute carrier family 38, member 7",SLC38A7,1.5,6.4e-05
You would specify this in your html file:
window.venn_settings = { csv_file: 'data.csv',
key_column: 'gene-list',
id_column: 'id'
info_columns: ['id', 'Description', 'Gene Name']
}
or using vennt.py
python vennt.py data.csv --key gene-list --id id --info id Description 'Gene Name'
Feel free to contribute with pull requests, bug reports or enhancement suggestions.
For building from sources, you will need nodejs and the following modules.
npm install -g browserify
npm install -g clean-css
npm install hbsfy@1.3
npm install handlebars-runtime
npm install coffeeify # Needs to be local?
# Builds files index.html, main.js, main.min.css into build/
./build.sh
This will watch the js & coffeescript files and rebuild main.js as needed. You'll still need to build the css using build.sh.
npm install -g watchify
watchify -t coffeeify -t hbsfy --debug app/main.coffee -o build/main.js -v
(cd build ; python -mSimpleHTTPServer)
Vennt is released under the GPL v3 (or later) license, see COPYING.txt