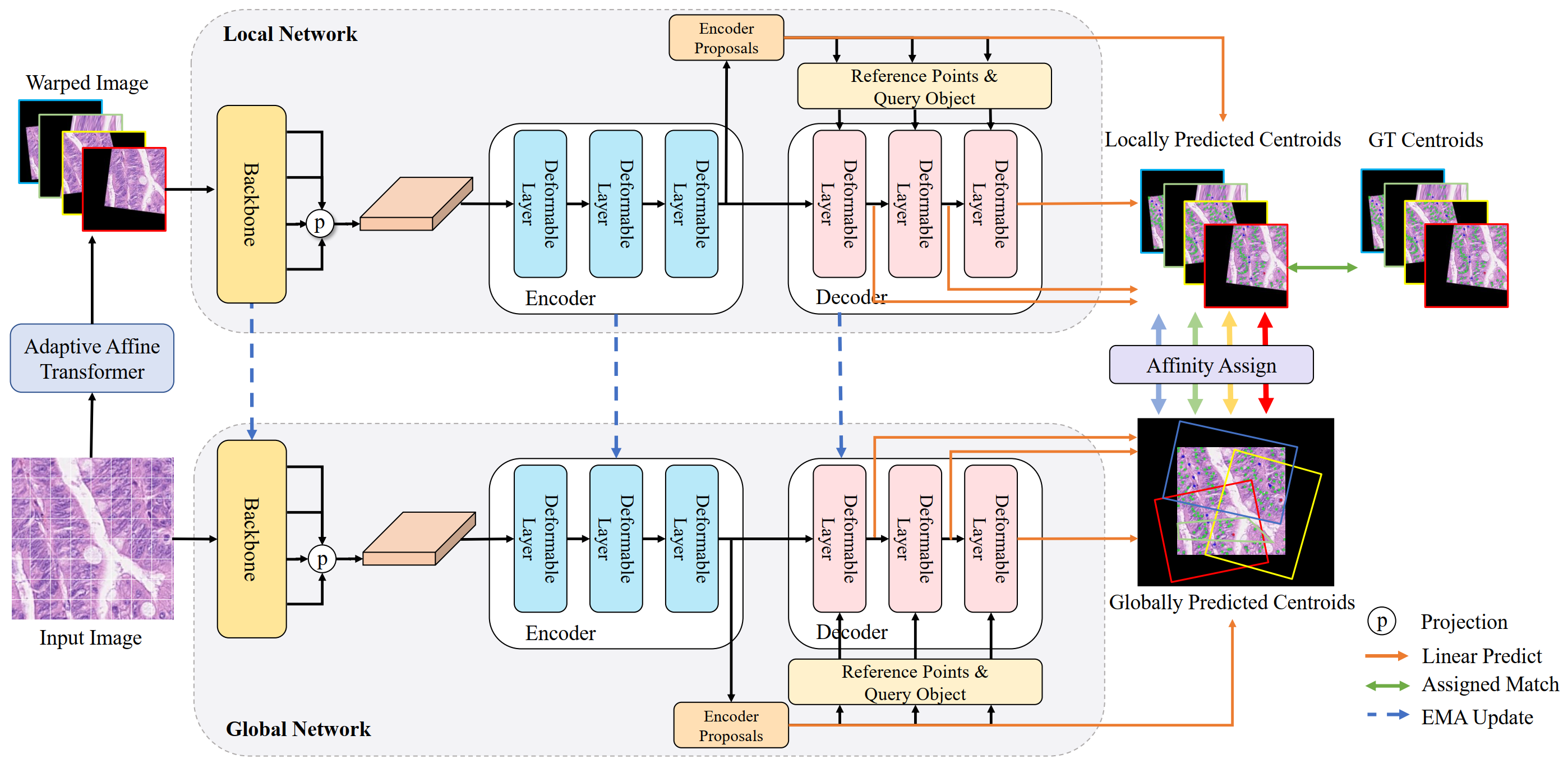
Code for Affine-Consistent Transformer for Multi-Class Cell Nuclei Detection (ICCV 2023)
(Continually updating ...)
-python=3.8
-pytorch=1.12.1+cu102
Install mmcv using mim
pip install -U openmim
mim install mmcv-full==1.6.1
Git clone acformer
git clone https://github.com/LL3RD/ACFormer.git
Install
cd ACFormer
cd thirdparty/mmdetection
python -m pip install -e .
cd ../..
python -m pip install -e .
Your can download Original Lizard from the official website or Preprocessed Lizard that is converted to hovernet consep format and split into patches.
Your can download 20x CoNSeP Dataset from here.
Your can download BRCA Dataset from the official website or Preprocessed BRCA.
| Method | F1d | F1c | Model Weights | Config Files |
|---|---|---|---|---|
| ACFormer | 0.782 | 0.557 | Checkpoint | Config |
| Method | F1d | F1c | Model Weights | Config Files |
|---|---|---|---|---|
| ACFormer | 0.739 | 0.613 | Checkpoint | Config |
| Method | F1d | F1c | Model Weights | Config Files |
|---|---|---|---|---|
| ACFormer | 0.796 | 0.485 | Checkpoint | Config |
First Download the preprocess dataset and change the dataset path in config/ACFormer_CoNSeP.py and Run
CUDA_VISIBLE_DEVICES=0 bash tools/dist_train.sh configs/ACFormer_CoNSeP.py 1 --work-dir=Path to save
First Install the sahi package.
cd tools/sahi
pip install -e .
cd ..
Then Prepare the slicing dataset. (Modify the CoNSeP Path in prepare_consep_dataset_40x.py)
python prepare_consep_dataset_40x.py
Change the dataset path in config/ACFormer_CoNSeP_40x.py and Run
CUDA_VISIBLE_DEVICES=0 bash tools/dist_train.sh configs/ACFormer_CoNSeP_40x.py 1 --work-dir=Path to save
Download the preprocessed dataset and Modify your dataset path and checkpoint path in tools/inference_lizard.py and run
python tools/inference_lizard.py
python tools/inference_consep.py
python tools/inference_brca.py
- ACFormer is built based on SoftTeacher and MMDetection.