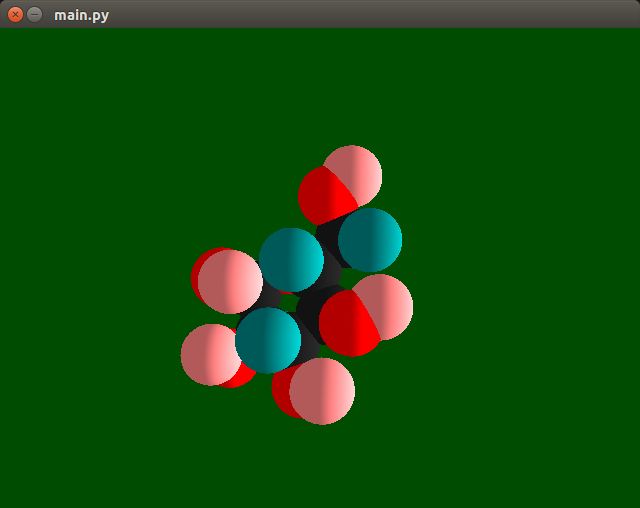
Molecular viewer
Draw atoms in different colors and place them in different places of space, thus obtaining a molecule. To do this, you need to know the coordinates, that's why chemical expert system Open Babel is used, so you need to install it as a prerequisite package on your system to run successfully following scripts:
apt-get install python-openbabel
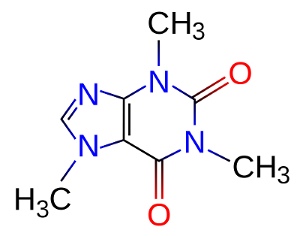
Using it, you can transform the SMILES formula to the list of atoms coordinates. SMILES can be found in Wikipedia articles about various substances.
For example, glucose: OC[C@H]1OC(O)[C@H](O)[C@@H](O)[C@@H]1O.
And its coordinates:
O3 3.08232699168 1.41136753836 1.97383867659
C3 2.63605783234 0.116724346362 2.37092125466
C3 2.87897272901 -0.854338624684 1.21070216538
H 2.47296741411 -0.36351143938 0.319191621385
O3 4.29331227545 -1.03408976253 1.03052197614
C3 2.18198168708 -2.2062426467 1.4123437219
H 2.52686439483 -2.70828022178 2.32628341407
O3 0.757046712076 -2.07375697965 1.48816465135
C3 2.4834103428 -3.09613160782 0.198042289604
H 2.03158506981 -2.66605342385 -0.704362586236
O3 1.84366255476 -4.36331585678 0.373985237387
C3 3.99532722341 -3.24522583591 0.00523555683618
H 4.41250004827 -3.77444425583 0.871020029013
O3 4.29626886035 -4.03252960027 -1.15195954921
C3 4.62954125722 -1.84273135567 -0.0947112115177
H 5.71887138656 -1.95252927548 -0.108137148969
O3 4.3079363412 -1.18316983652 -1.31789119536
HO 2.91473185309 2.02688340774 2.71060639162
H 1.56956736943 0.19992674022 2.5985040442
H 3.17829013287 -0.18365904297 3.27321534863
HO 0.532102901531 -1.64213847492 2.33074495499
HO 0.907383263524 -4.15051515566 0.55676359202
HO 3.89297302828 -3.59188323274 -1.91963488602
HO 3.3506978517 -1.05986229308 -1.37211748251
You can use module render.py to visualize Python code. It is an entry point to run viewer app based on OpenGL library.
MIT licensed library. See LICENSE for details.
If you have suggestions for improving the pymolecule, please open an issue or pull request on GitHub.