You need to either create an environment or update an existing environment. After creating an environment you have to activate it:
conda activate pyPDPPartitioner
conda env create -f environment.yml
conda env update -f environment.yml --prune
pip install pyPDPPartitioner
For HPO-Bench examples, you further need to install HPOBench from git (e.g. pip install git+https://github.com/automl/HPOBench.git@master).
To use this package you need
- A Blackbox function (a function that gets any input and outputs a score)
- A Configuration Space that matches the required input of the blackbox function
There are some synthetic Blackbox-functions implemented that are ready to use:
f = StyblinskiTang.for_n_dimensions(3) # Create 3D-StyblinskiTang function
cs = f.config_space # A config space that is suitable for this functionTo sample points for fitting a surrogate, there are multiple samplers available:
- RandomSampler
- GridSampler
- BayesianOptimizationSampler with Acquisition-Functions:
- LowerConfidenceBound
- (ExpectedImprovement)
- (ProbabilityOfImprovement)
sampler = BayesianOptimizationSampler(f, cs)
sampler.sample(80)All algorithms require a SurrogateModel, which can be fitted with SurrogateModel.fit(X, y) and yields means and variances with SurrogateModel.predict(X).
Currently, there is only a GaussianProcessSurrogate available.
surrogate = GaussianProcessSurrogate()
surrogate.fit(sampler.X, sampler.y)There are some available algorithms:
- ICE
- PDP
- DecisionTreePartitioner
- RandomForestPartitioner
Each algorithm needs:
- A
SurrogateModel - One or many selected hyperparameter
- samples
num_grid_points_per_axis
Samples can be randomly generated via
# Algorithm.from_random_points(...)
ice = ICE.from_random_points(surrogate, selected_hyperparameter="x1")Also, all other algorithms can be built from an ICE-Instance.
pdp = PDP.from_ICE(ice)
dt_partitioner = DecisionTreePartitioner.from_ICE(ice)
rf_partitioner = RandomForestPartitioner.from_ICE(ice)The Partitioners can split the Hyperparameterspace of not selected Hyperparameters into multiple regions. The best region can be obtained using the incumbent of the sampler.
incumbent_config = sampler.incumbent_config
dt_partitioner.partition(max_depth=3)
dt_region = dt_partitioner.get_incumbent_region(incumbent_config)
rf_partitioner.partition(max_depth=1, num_trees=10)
rf_region = rf_partitioner.get_incumbent_region(incumbent_config)Finally, a new PDP can be obtained from the region. This PDP has the properties of a single ICE-Curve since the mean of the ICE-Curve results in a new ICE-Curve.
pdp_region = region.pdp_as_ice_curveMost components can create plots. These plots can be drawn on a given axis or are drawn on plt.gca() by default.
sampler.plot() # Plots all samplessurrogate.plot_means() # Plots mean predictions of surrogate
surrogate.plot_confidences() # Plots confidencessurrogate.acq_func.plot() # Plot acquisition function of surrogate modelice.plot() # Plots all ice curves. Only possible for 1 selected hyperparameterice_curve = ice[0] # Get first ice curve
ice_curve.plot_values() # Plot values of ice curve
ice_curve.plot_confidences() # Plot confidences of ice curve
ice_curve.plot_incumbent() # Plot position of smallest value pdp.plot_values() # Plot values of pdp
pdp.plot_confidences() # Plot confidences of pdp
pdp.plot_incumbent() # Plot position of smallest value dt_partitioner.plot() # only 1 selected hp, plots all ice curves in different color per region
dt_partitioner.plot_incumbent_cs(incumbent_config) # plot config space of best region
rf_partitioner.plot_incumbent_cs(incumbent_config) # plot incumbent config of all treesregion.plot_values() # plot pdp of region
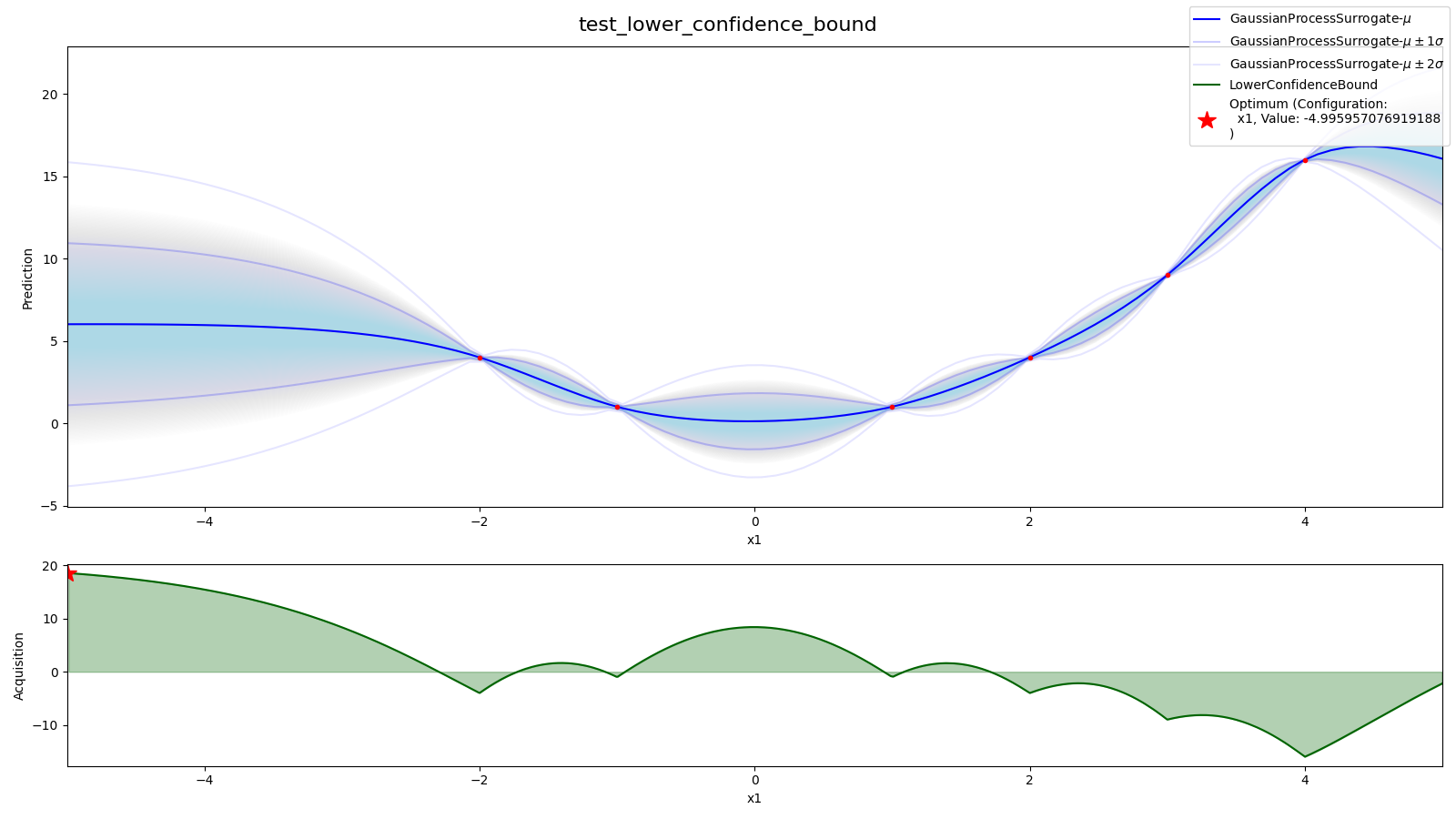
region.plot_confidences() # plot confidence of pdp in regionSource: tests/sampler/test_acquisition_function.py
- 1D-Surrogate model with mean + confidence
- acquisition function
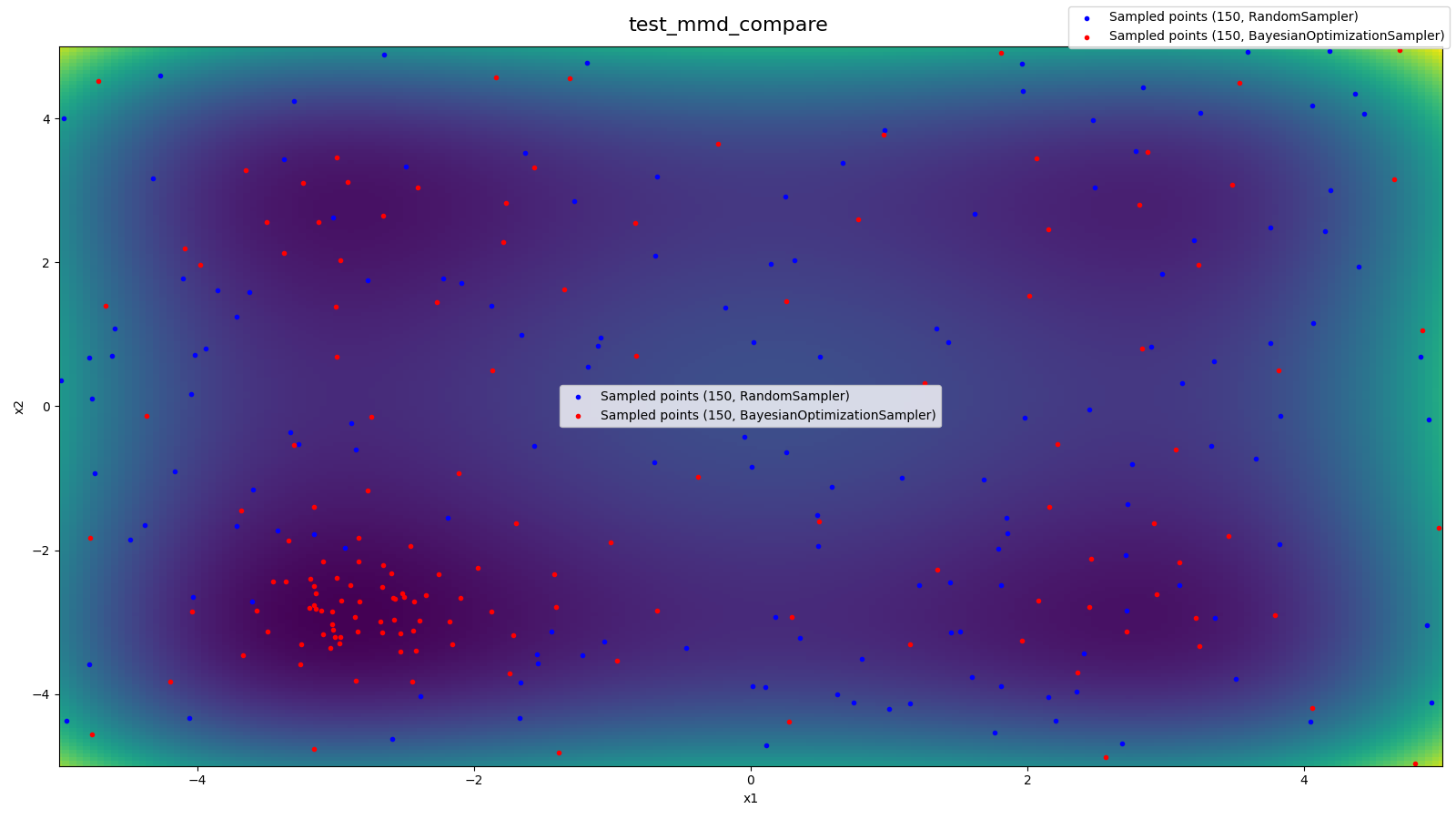
Source: tests/sampler/test_mmd.py
- Underlying blackbox function (2D-Styblinski-Tang)
- Samples from RandomSampler
- Samples from BayesianOptimizationSampler
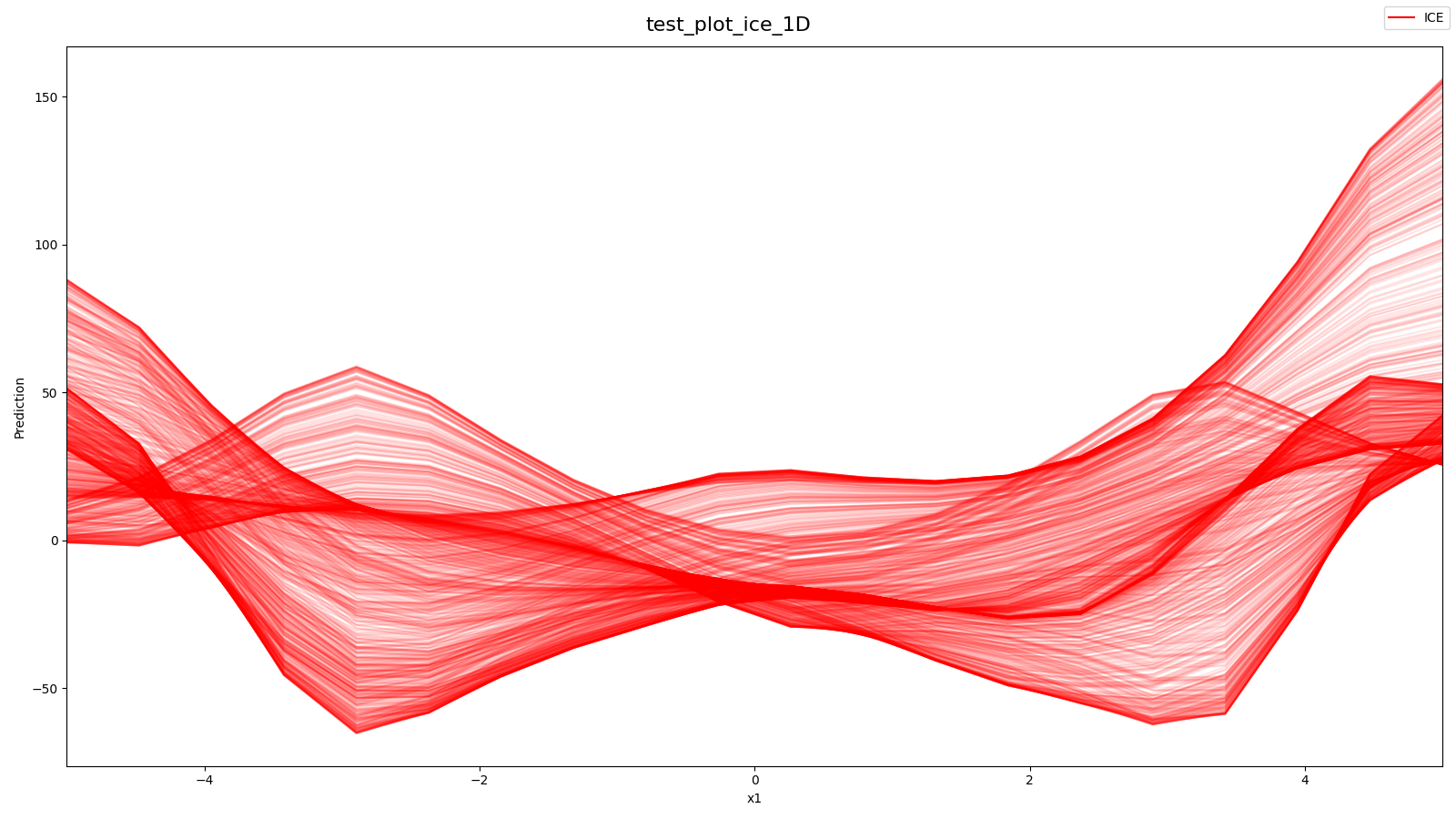
Source: tests/algorithms/test_ice.py
- All ICE-Curves from 2D-Styblinski-Tang with 1 selected Hyperparameter
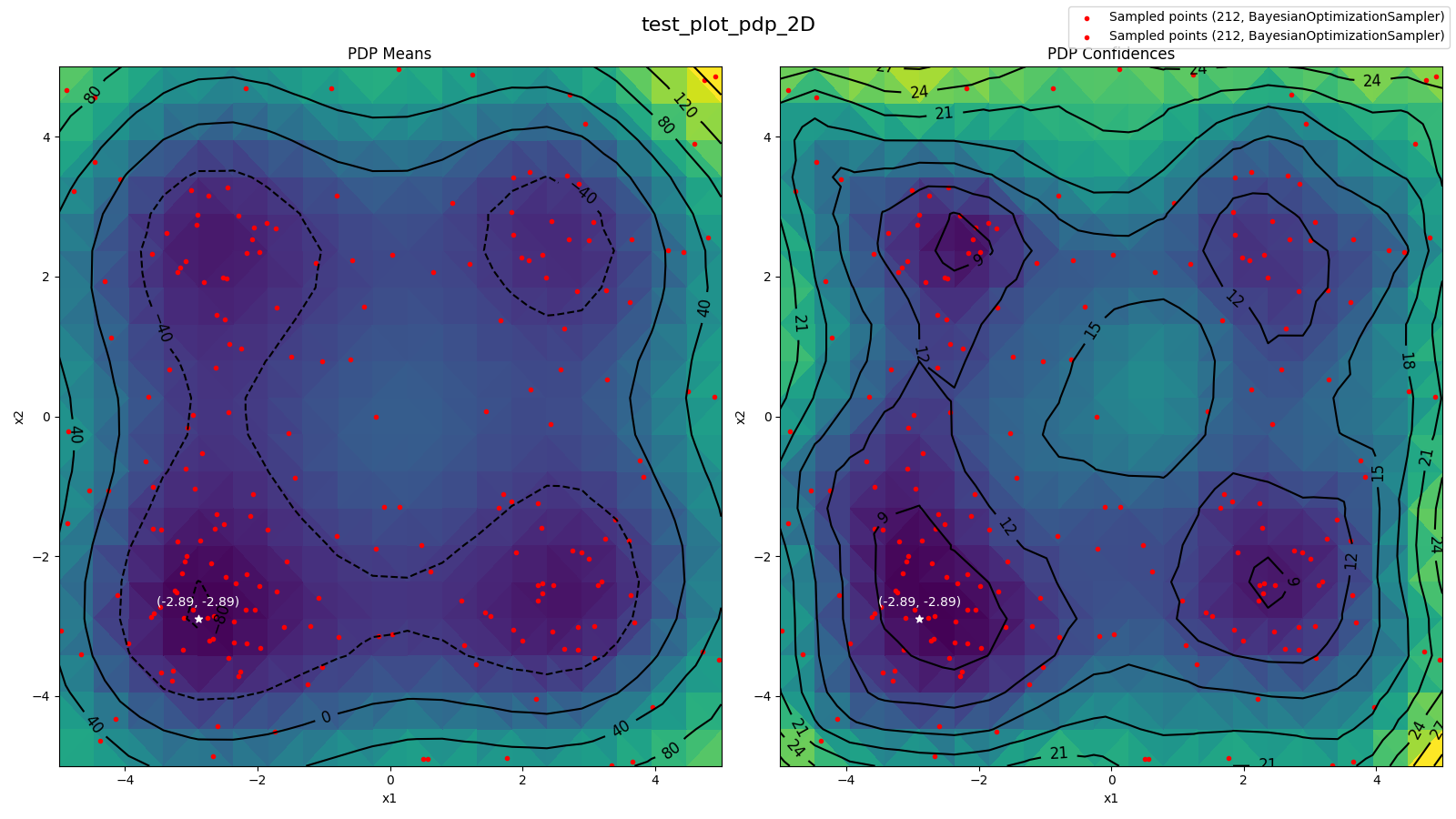
Source: tests/algorithms/test_pdp.py
- 2D PDP (means)
- 2D PDP (confidences)
- All Samples for surrogate model
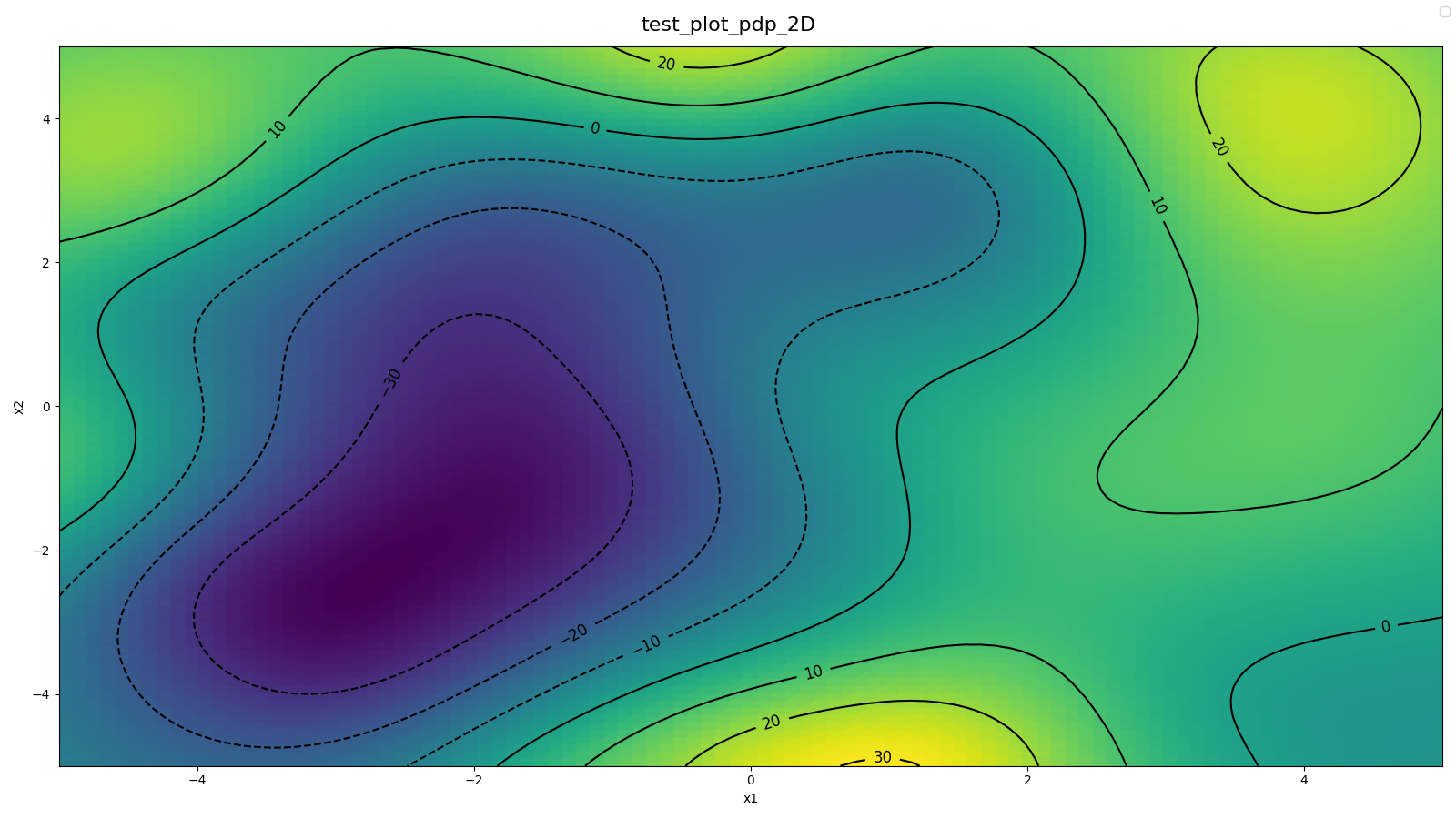
Source: examples/main_2d_pdp.py (num_grid_points_per_axis=100)
- 2D PDP (means)
Source: tests/algorithms/partitioner/test_partitioner.py
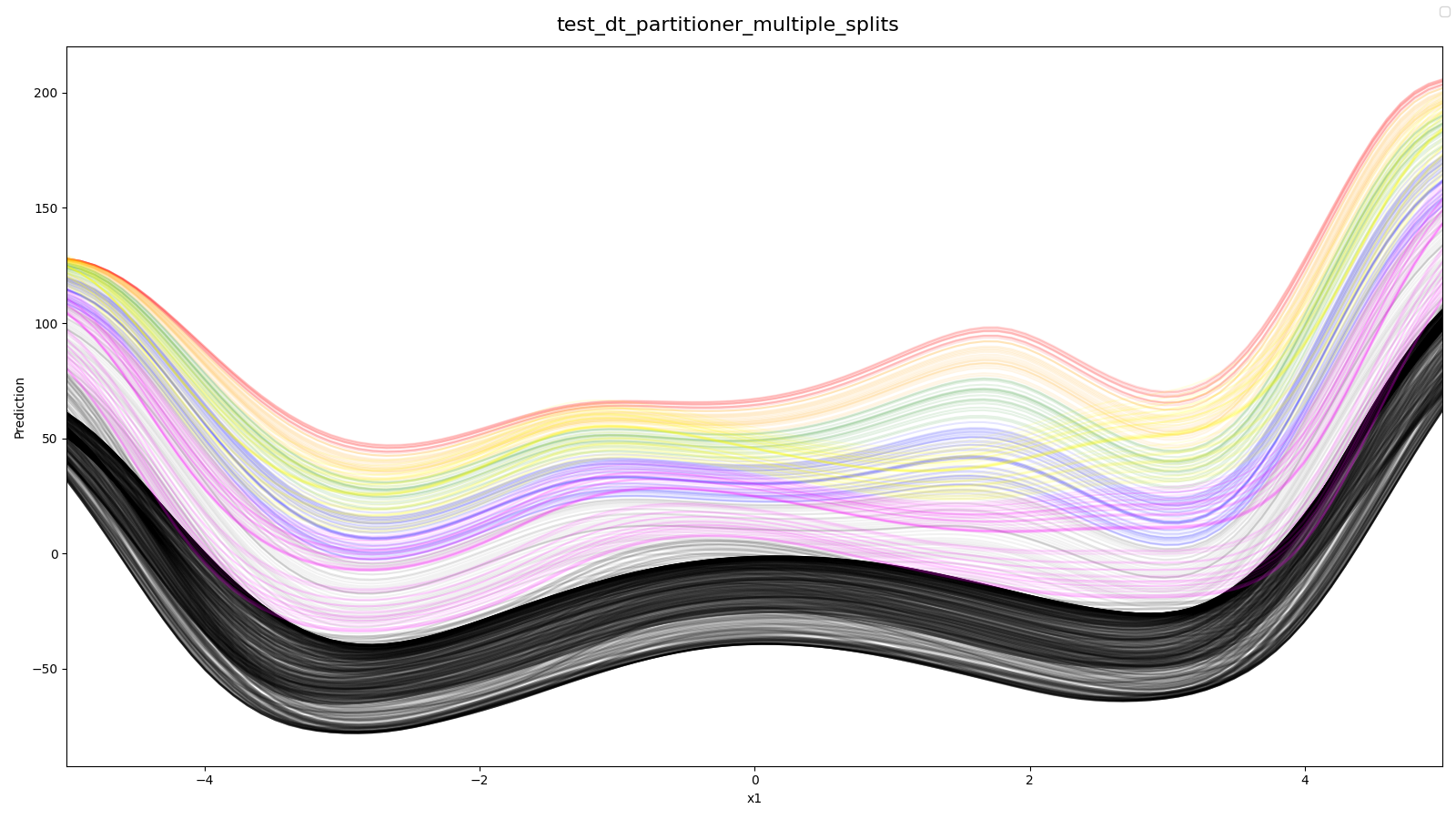
- All ICE-Curves splitt into 8 different regions (3 splits) (used 2D-Styblinski-Tang with 1 selected hyperparameter)
Source: tests/algorithms/partitioner/test_partitioner.py
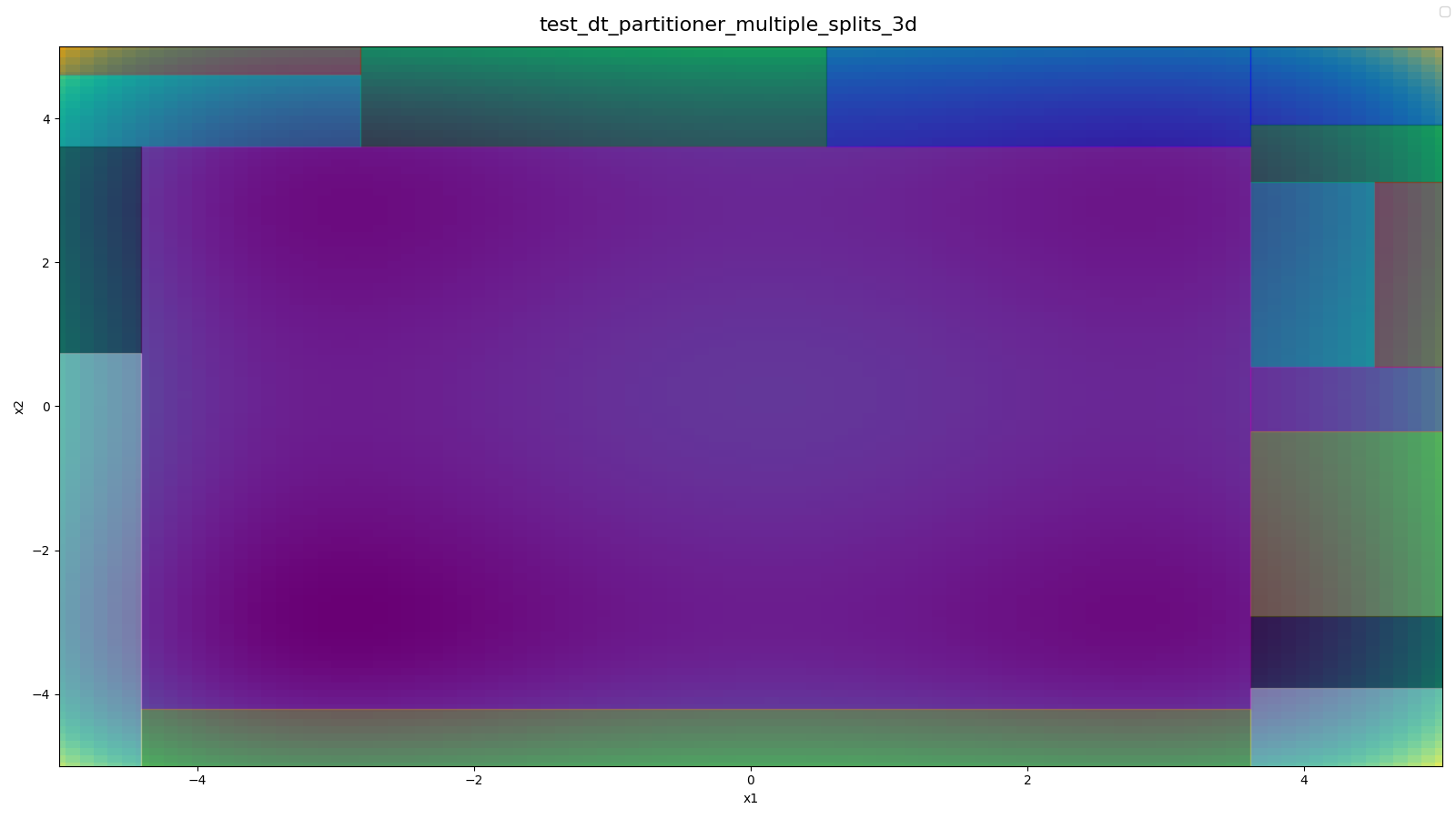
- All Leaf-Config spaces from Decision Tree Partitioner with 3D-Styblinski-Tang Function and 1 Selected Hyperparameter (
x3) - 2D-Styblinkski-Tang in background