DeepDecon: A Deep-learning Method for Estimating Cell Fractions in Bulk RNA-seq Data with Applications to AML
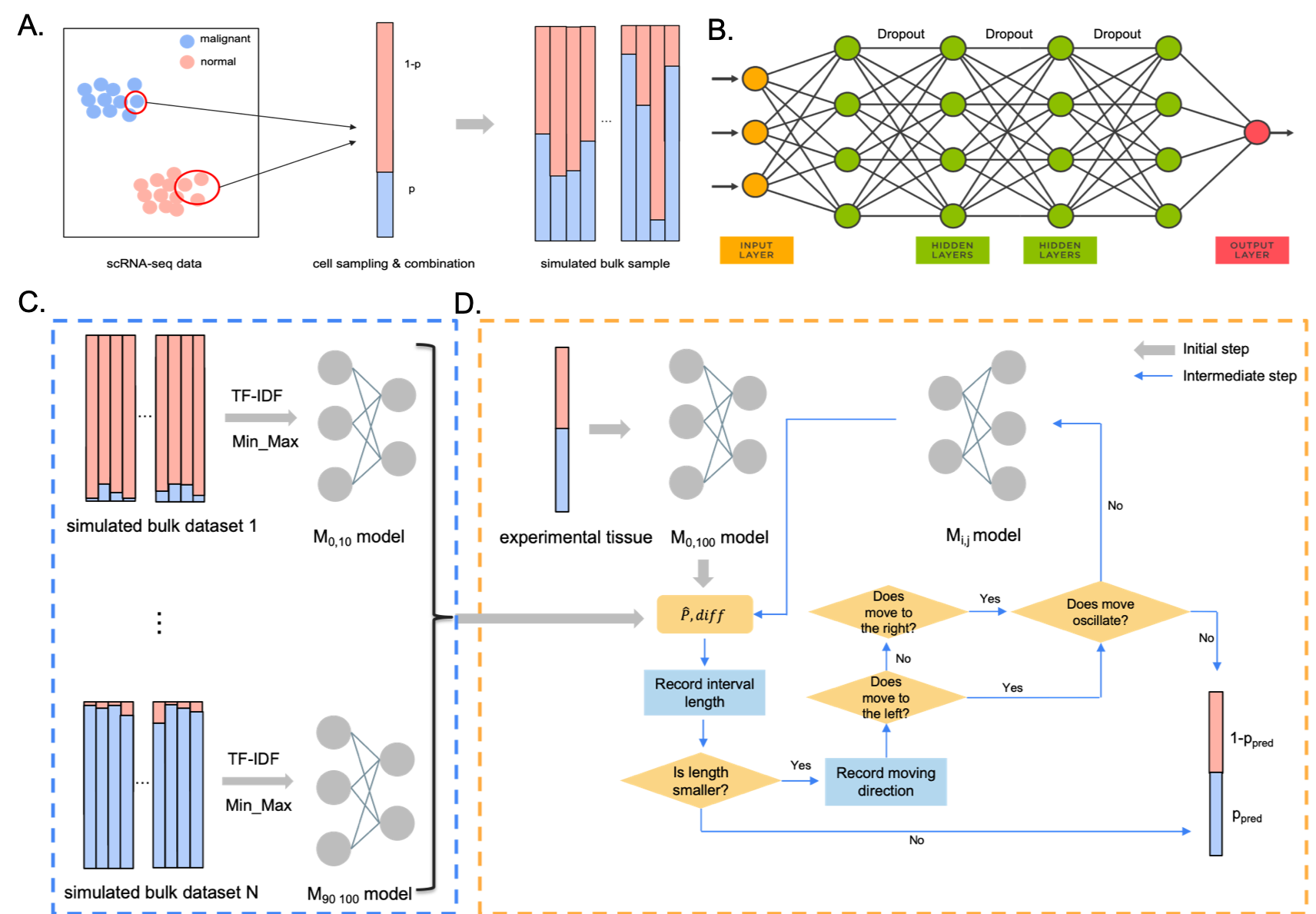
Here, we present DeepDecon, a deep neural network model leveraging single-cell gene expression information to accurately predict the fraction of cancer cells in bulk tissues. DeepDecon was trained based on single-cell RNA sequencing data and was robust to experimental biases and noises. It will automatically select optimal models to recursively estimate malignant cell fractions and improve prediction accuracy. When applied to bone marrow data (see Tutorials), it outperforms existing decomposition methods in both accuracy and robustness. We further show that the DeepDecon is robust to the number of single cells within a bulk sample.
- tensorflow 1.14.0
- scikit-learn 0.24.2
- python 3.6.12
- pandas 1.1.3
- numpy 1.19.2
- keras 2.3.1
- scanpy 1.7.2
Download DeepDecon by
git clone https://github.com/Jiawei-Huang/DeepDecon.git
Installation has been tested in a Linux and MacOs platform with Python3.6. GPU is recommended for accelerating the training process.
This section provides instructions on how to run DeepDecon with scRNA-seq datasets.
Several scRNA-seq AML datasets have been prepared as the input of DeepDecom model. These datasets can be downloaded from the zenode repository. Uncompress the datasets.tar.gz in datasets folder then each dataset will have its own file, which denotes the gene expression matrix (XXX_norm_sc.txt, XXX refers to the subject name). Each row in the matrix refers to one cell and the first column of the matrix refers to the cell type (malignant/normal), the rest columns refer to genes.
DeepDecon construct bulk RNA-seq samples through the get_bulk_samples.py script. One can try generate a bulk RNA-seq dataset with any ratio of malignant cell by running
python ./src/get_bulk_samples.py [-h] [--cells CELLS] [--samples SAMPLES] [--subject SUBJECT] [--start START] [--end END] [--binomial BINOMIAL] [--data DATA] [--out OUT]
-h, --help show this help message and exit
--cells CELLS Number of cells to use for each bulk sample.
--samples SAMPLES, -n SAMPLES
Total number of samples to create for each dataset.
--subject SUBJECT Subject name
--start START Fraction start range of generated samples e.g. 0 for [0, 100]
--end END Fraction end range of generated samples e.g. 0 for [0, 100]
--binomial BINOMIAL Whether generating bulk fractions from binomial distribution, 0=False, 1=True
--data DATA Directory containg the datsets
--out OUT Output directoryAs long as we have the data, one can train DeepDecon models by running
python train_model.py [-h] [--cells CELLS] [--path PATH] [--lr LR] [--bs BS]
[--dr DR] [--start START] [--end END] [--scaler SCALER]
[--normalization NORMALIZATION]
-h, --help show this help message and exit
--cells CELLS Number of cells to use for each bulk sample.
--path PATH Training data directory
--lr LR learning rate index k, lr = 10^(-k)
--bs BS batch size
--dr DR dropout
--start START Fraction start range of generated samples e.g. 0 for
[0, 100]
--end END Fraction end range of generated samples e.g. 100 for
[0, 100]
--scaler SCALER Scaler of neural network, MinMaxScaler (mms) or
StandardScaler (ss)
--normalization NORMALIZATION
Normalization methods,TF-IDF, FPKM, CPM or TPMNext, people can get predictions by running
python eval.py [--cells CELLS] [--dir DIR] [--filepath FILEPATH] [--sub_idx SUB_IDX]
--cells CELLS Number of cells to use for each bulk sample.
--dir DIR Training data directory
--filepath FILEPATH Testing file path
--sub_idx SUB_IDX Testing subject index, 0-14 refers to subjects in the
training datasets, 15 means new dataset.See DeepDecon_example.ipynb for reproducing the experimental results in this paper.
Feel free to open an issue on Github or contact me if you have any problem in running DeepDecon.