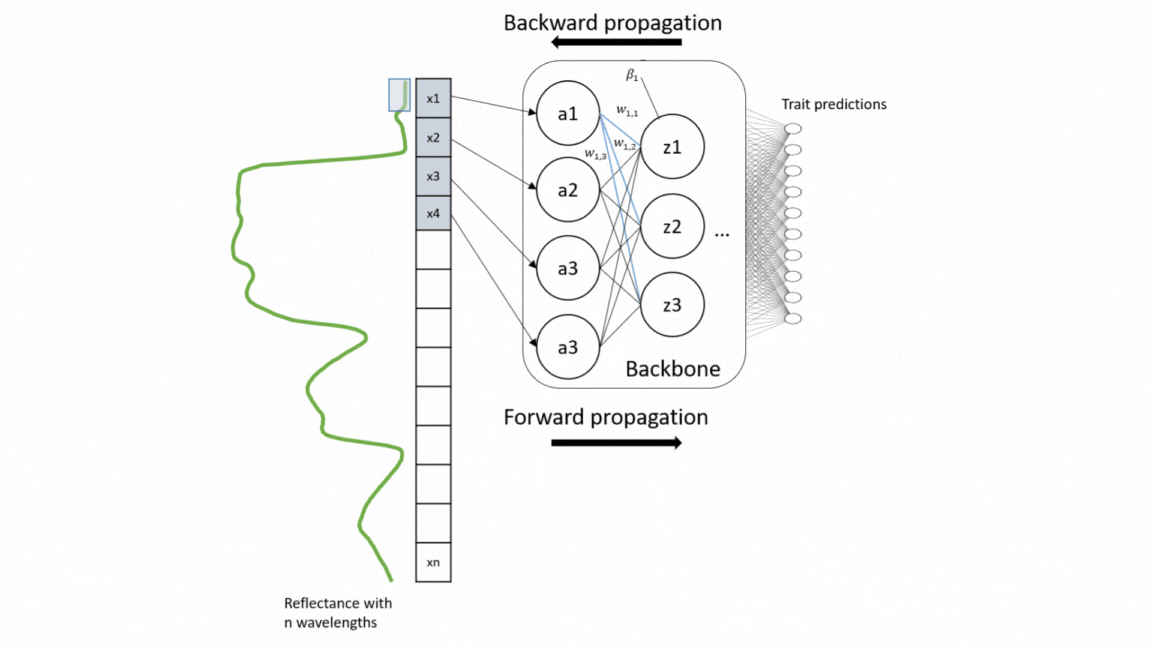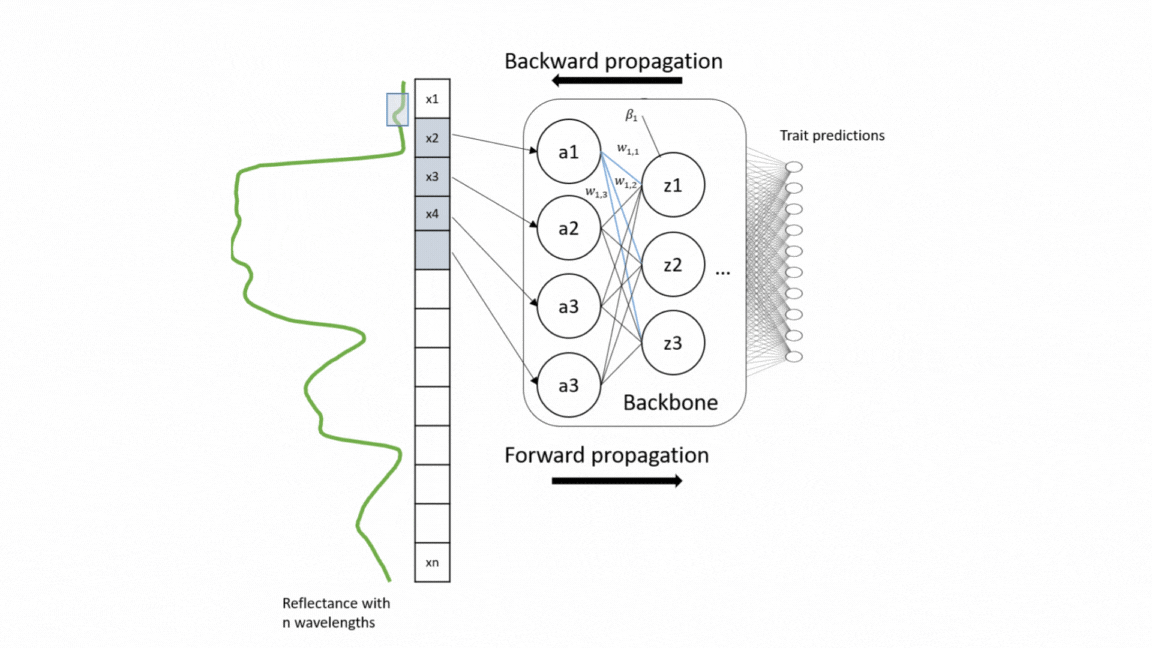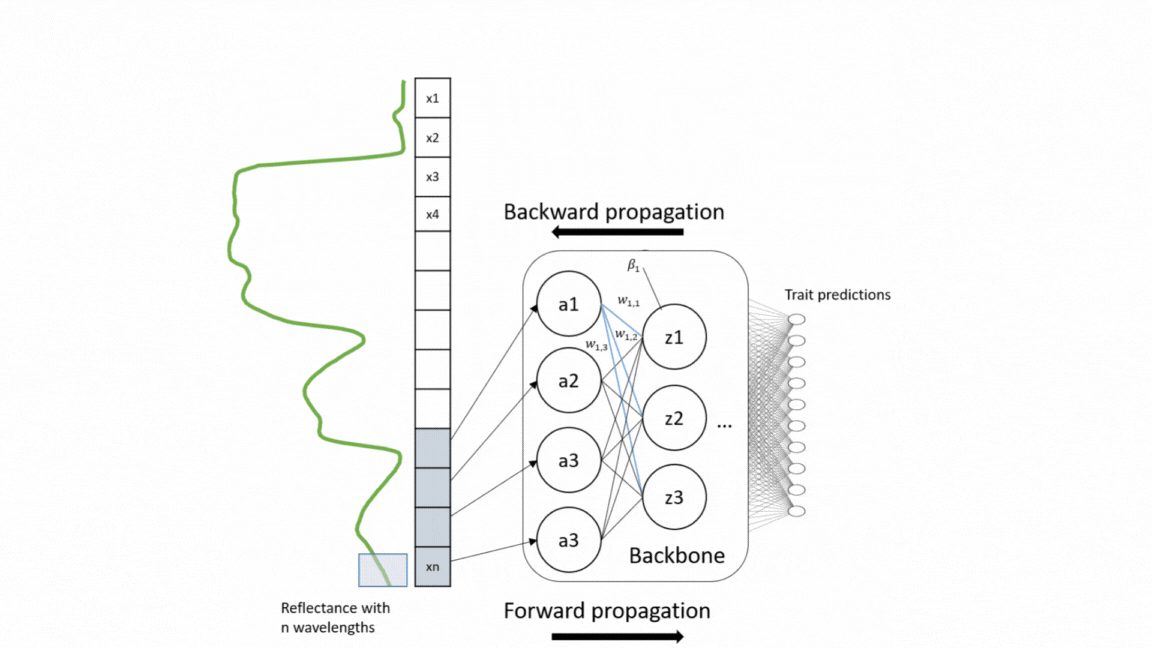
For a wide range of applications, including vegetation modeling in Earth systems models, nature conservation, and forest monitoring, global information on functional plant traits is essential. Yet the coverage of concurrent measurement of multiple plant traits across different ecosystem types is still sparse. With the upcoming unprecedented amount of spectroscopy data, we present here a model that simultaneously retrieve 20 structural and biochemical traits from canopy reflectance data (Multi-trait model).
This repository is built upon models developed by Cherif et al. 2023_RSE For the model training, a large number of data sets of canopy spectra and their corresponding leaf trait measurements were compiled, including 42 data sets from different ecosystem types and sesnor types. The multi-trait model was developed on a weakly supervised learning approach and therefore trained on this heterogeneous and sparse data. The used architecture of the model was an adapted version of EfficientNetB0 based on 1D-CNN. This architecture enables to extract interrelationships from the reflectance data as well as among traits. For validation, the model was evaluated on different data sets as external validation (transferability) and compared with the widely-used Partial Least Square Regression (PLSR) models as well as single CNN models. For in-depth technical insights into the model development process, please refer to the following repository: code_multi.
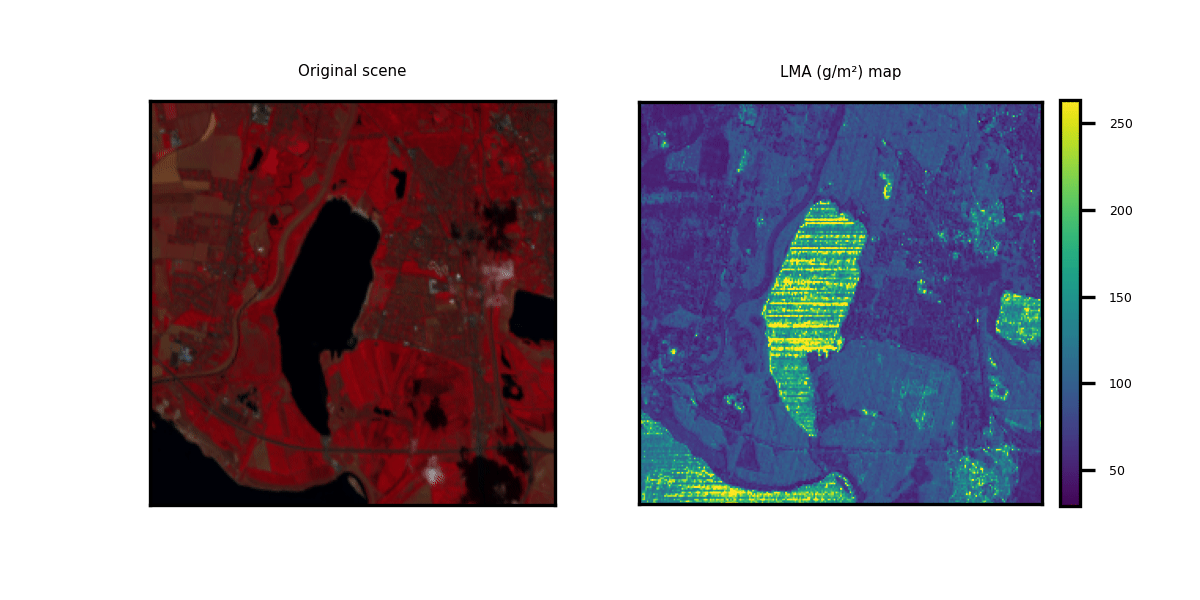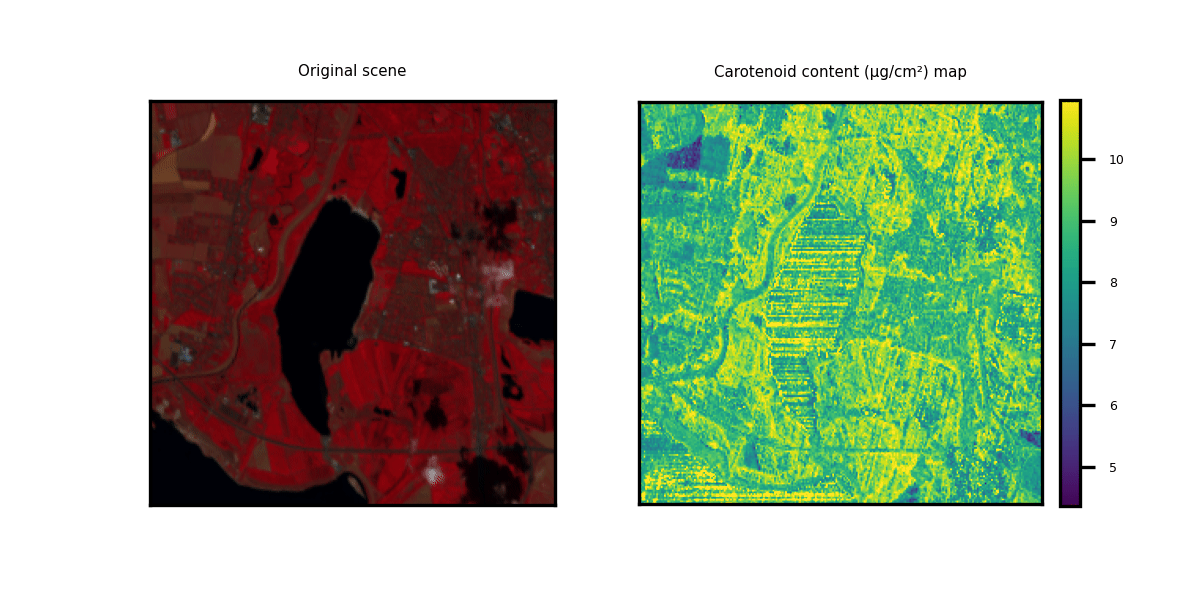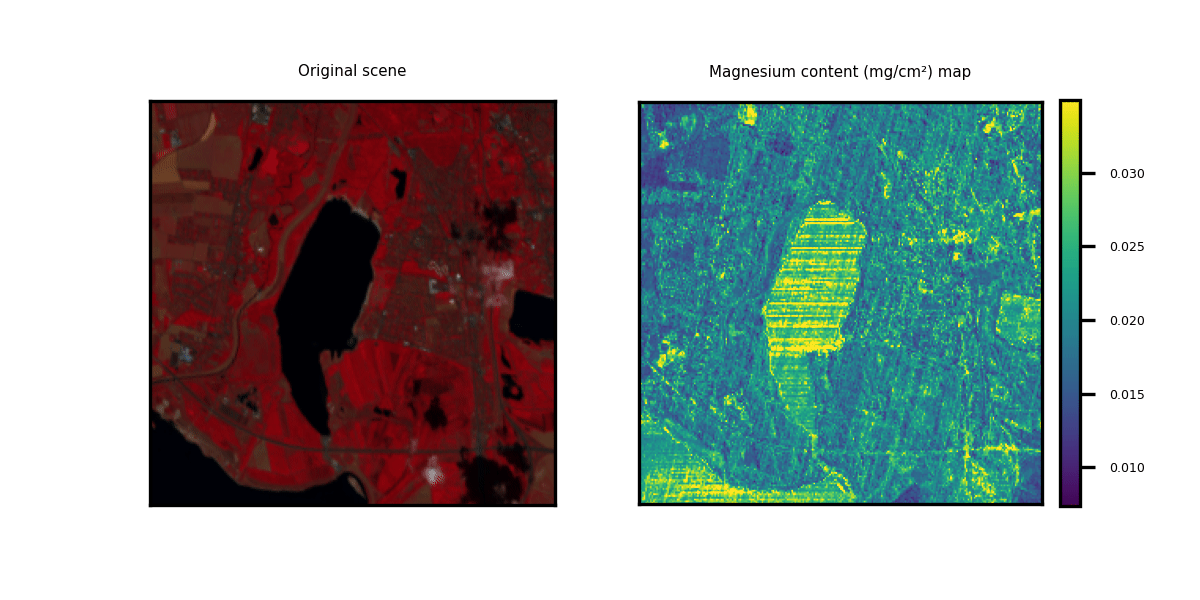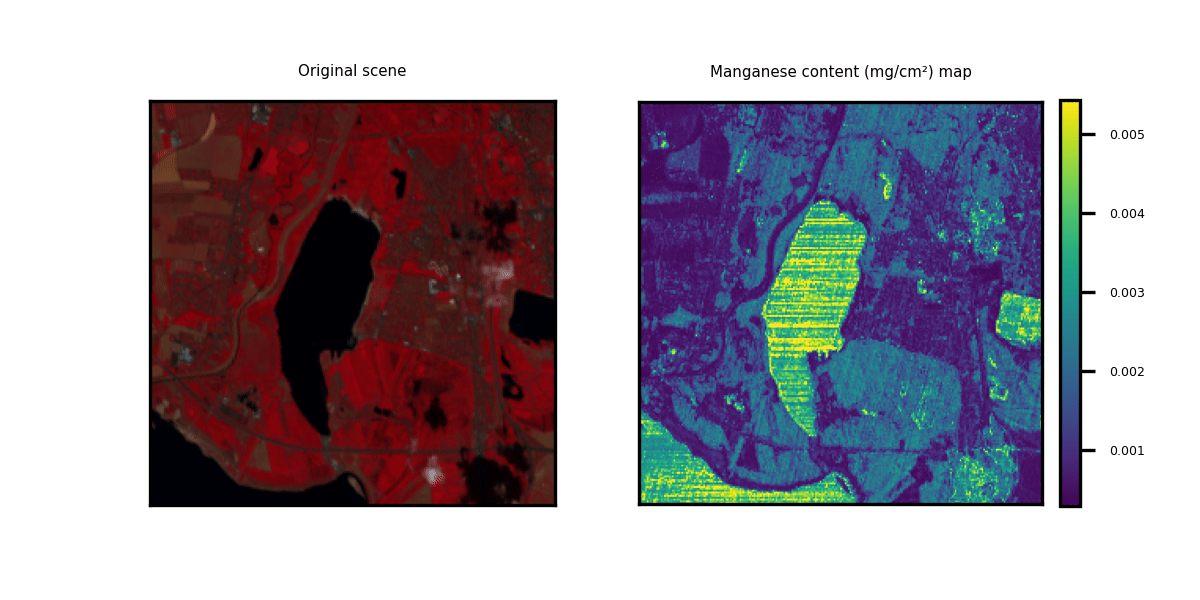
This repository showcases practical applications through hands-on examples for generating multi-trait maps from Hyperspectral Imagery (HSI). While two HSI examples are provided for testing purposes, users have the flexibility to upload new scenes along with relevant information about the available bands.
We provide three options to deploy our model on new hyperspectral scenes in GeoTIFF format. The data can be of any shape (height and width) but the channels should correspond to the bands indicated in the .csv metadata file. Two toy examples are provided.
- Option_1: Clone the repository and run the inference script
This project is based on tensorflow v2.7.0 and python v3.9.5.
- Clone this repository
conda create -n <environment-name> python==3.9conda activate <environment-name>- Install tensorflow (this model was tested for =2.7.0). May vary with your system.
cdinto the cloned repopip install -r requirements.txt- Run this command to test on a Toy scene (default enmap_toyExp.tif)
python HS_predictions.py --sceneText ToyEnmap
- Option_2: For a smoother execution of the code, we have also made available this Colab notebook
- Option_3: Or use our interactive Demo at
If you have data, please contact us, it would be nice to have you on board for future studies! We are committed to continually enhancing the model's generalizability, and your contributions play a crucial role.
Found a bug or encountering difficulties while running the models? Your feedback is invaluable! Please reach out to us with details about any issues you encounter. Your insights help us refine and improve the overall performance.
- The Python-Scripts and notebooks for model devlopment can be found in code_multi.
- The updated model object can found in models directory. As we are constantly improving the model with new data, please make sure to get the last version.
- EGU23 contribution Session BG9.4
If this repository helped your research, please cite Cherif et al. RSE, 2023. Here is an example BibTeX entry:
@article{CHERIF2023113580,
title = {From spectra to plant functional traits: Transferable multi-trait models from heterogeneous and sparse data},
journal = {Remote Sensing of Environment},
volume = {292},
pages = {113580},
year = {2023},
issn = {0034-4257},
doi = {https://doi.org/10.1016/j.rse.2023.113580},
url = {https://www.sciencedirect.com/science/article/pii/S0034425723001311},
author = {Eya Cherif and Hannes Feilhauer and Katja Berger and Phuong D. Dao and Michael Ewald and Tobias B. Hank and Yuhong He and Kyle R. Kovach and Bing Lu and Philip A. Townsend and Teja Kattenborn},
keywords = {Hyperspectral remote sensing, Plant trait retrieval, Deep learning, Biophysical variables, Imaging spectroscopy, Canopy properties, Weakly supervised learning, Multi-task regression},