REddyProc package supports processing (half)hourly data from Eddy-Covariance sensors.
There is an online-formular to use the functionality of the package including description at https://www.bgc-jena.mpg.de/bgi/index.php/Services/REddyProcWeb.
# Release stable version from CRAN
install.packages("REddyProc")
# The development version from GitHub using devtools:
# install.packages("devtools")
devtools::install_github("bgctw/REddyProc")A simple example performs Lookuptable-based gapFilling of Net-Ecosystem-Exchange (NEE) and plotting a fingerprint plot of the filled values.
library(REddyProc)
#+++ Input data from csv (example needs to be downloaded)
examplePath <- getExamplePath('Example_DETha98.txt', isTryDownload = TRUE)
if (length(examplePath)) {
EddyData.F <- fLoadTXTIntoDataframe(examplePath)
} else {
warning(
"Could not find example text data file."
," In order to execute this example code,"
," please, allow downloading it from github. "
," Type '?getExamplePath' for more information.")
# using RData version distributed with the package instead
EddyData.F <- Example_DETha98
}
#+++ If not provided, calculate VPD from Tair and rH
EddyData.F$VPD <- fCalcVPDfromRHandTair(EddyData.F$rH, EddyData.F$Tair)
#+++ Add time stamp in POSIX time format
EddyDataWithPosix.F <- fConvertTimeToPosix(
EddyData.F, 'YDH', Year.s = 'Year', Day.s = 'DoY', Hour.s = 'Hour')
#+++ Initalize R5 reference class sEddyProc for processing of eddy data
#+++ with all variables needed for processing later
EddyProc.C <- sEddyProc$new(
'DE-Tha', EddyDataWithPosix.F, c('NEE','Rg','Tair','VPD', 'Ustar'))
#Location of DE-Tharandt
EddyProc.C$sSetLocationInfo(
Lat_deg.n = 51.0, Long_deg.n = 13.6, TimeZone_h.n = 1)
#
#++ Fill NEE gaps with MDS gap filling algorithm (without prior ustar filtering)
EddyProc.C$sMDSGapFill('NEE', FillAll.b = FALSE)
#
#++ Export gap filled and partitioned data to standard data frame
FilledEddyData.F <- EddyProc.C$sExportResults()
#
#++ Example plots of filled data to screen or to directory \plots
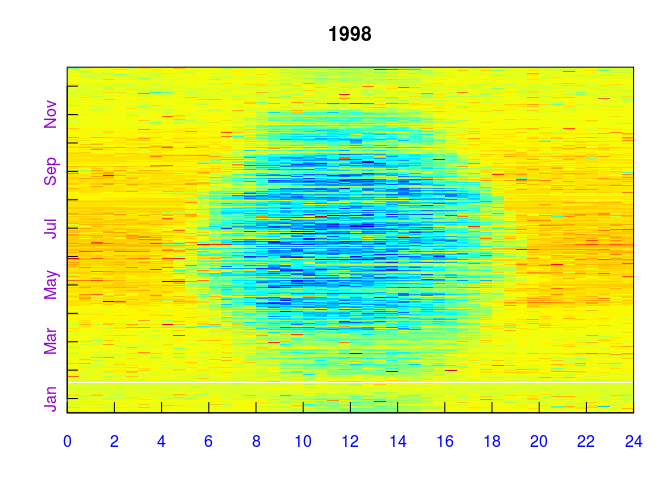
EddyProc.C$sPlotFingerprintY('NEE_f', Year.i = 1998)Further examples are in vignette(useCase) and vignette(DEGEbExample)