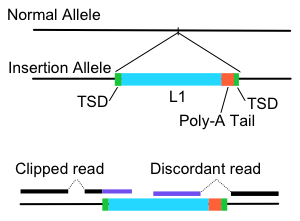
TE-detective identifies transposable element (TE) insertions in short read sequecing data such as produced by Illumina sequencers. We have benchmarked it using data from both mouse and human genomes.
Requirements
Installation
Docker Image
Definitions
Input Files
General Analysis Workflow
Results Files
Simple Example
Polymorphic Subtraction Example
Detailed Usage For Each Command
License Information for Externals
-
Python Packages
- numpy
- pysam
- biopython
Installation files for NCBI blast and Censor can be found in the externals directory. We recommend using Anaconda to install the relevant python packages.
Clone and install with pip:
git clone https://github.com/edwardslab-wustl/TE-detective.git
cd TE-detective
pip install .
Be sure the following are added to your PATH:
/opt/anaconda3/bin/
/usr/local/bwa
/usr/local/blast-2.2.26/bin
Set the environmental variable:
BLASTDIR=/usr/local/blast-2.2.26/bin
preferably prepared using following alignment command:
bwa mem -M -Y -R $RG_LINE ref.fa test_1.fq test_2.fq | samtools view -b -S - > test_ref.bam
You can pre-index the file with samtools, or the bam file will be indexed in the preprocess script, if it hasn't been already.
Space- or tab-delimited file specifying the repeat to be examined (e.g. LINE) and location of a fasta file with its corresponding reference sequences: The first field is the repeat name. The second field is a fasta file containing the reference sequences for the repeat element. We recomend specifying the full path, but if the file can't be found the code will then search the directory the ref_fofn file is in as well as the current working directory for the fasta file. Reference sequences of repeat elements can be obtained from Repbase or other resources. See example file ref_fofn in the example_data folder.
Be sure to use the appropriate file with coordinates corresponding to the genome version used for alignment.
You can download the repeatmasker track data from the UCSC Genome Browser and filter with something like:
zcat rmsk.txt.gz | awk '{print $6"\t"$7"\t"$8"\t"$12;}' > rmsk_hg19.bed
Clipped read: Sequencing read where part only part maps to the genome. The other part could either not map to the genome or map else where.
Discordant read: Paired sequencing reads from the same DNA fragment where one read maps to the genome and the other read either maps to another region of the genome or in the wrong orientation. I.e. reads that are not in proper paired alignments.
Clipped and discordant reads could come from a TE insertion, a genomic rearrangement or alteration, or from various sequencing or library construction artifacts.
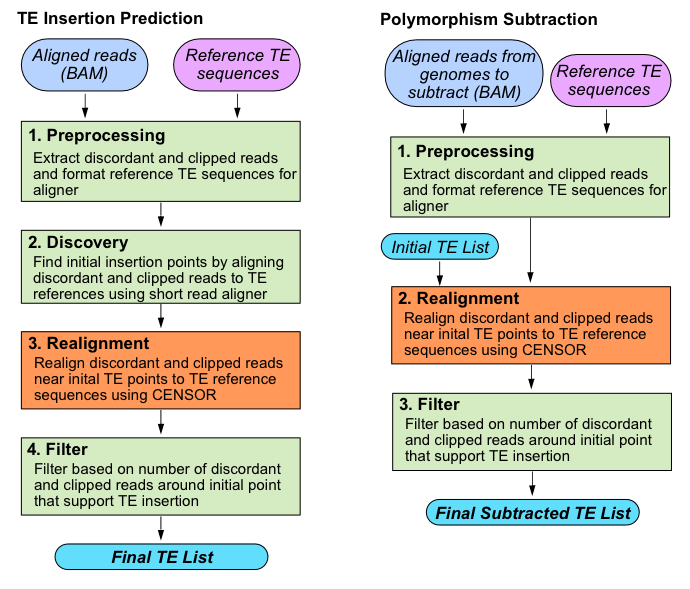
- TE_detective preprocess: extracts discordant and clipped reads from the bam file, sets up other files needed for the analysis
- TE_detective discover: determines an initial set of predictions that have discordant and clipped read support greater than the --discord_cluster_dens
- TE_detective nadiscover: performs some initial alignments, and determines poly A/T information
- TE_detective analyze: performs realignment around each initial insertion prediction. Calculates the number of discordant, clipped, and poly A/T reads.
- TE_detective filter: filters the initial set of predictions based on read support, existing TEs, polymorphic subtraction, or TE insertions predictions from another sample
- TE_detective cluster2d: optional module to quickly repreform the discover step using a different cutoff for read support with the --discord_cluster_dens parameter
- Perform general analysis workflow on each sample
- Run TE_detective analyze using the insertion predictions from the child, but specififying the preprocessing directories for the parent (run for each parent). This identifies potential support for an insertion in the parent at the predicted insertion points in the child.
- Run TE_detective filter specifying to filter based on the output(s) from the prior step.
These are the default file names. Output file names can be changed by the user.
List of initial TE insertion predictions after the discover step, along with a value for the initial amount of clipped and discordant read support found
List of revised initial TE insertion predictions after the nadiscover step
Revised list of initial TE insertion predictions after the cluster2D step
Results of realignment during the analyze step, this file will have an entry for every initial prediction, along with detailed information concerning the support for each insertion. This result is prior to any filtering. See the column headers for details.
The final filtered output from the filter step. See column headers for details.
Shows which filter each initial prediction failed or passed in the filter step
Basic stats on how many insertion predictions passed each filter in the filter step
tar -xzf TEdetective_example.tar.gz
cd TEdetective_example/example_data
Then either:
sh run_example.sh
or
TE_detective preprocess -i test_sim.bam -r ref_fofn
TE_detective discover -i test_sim.bam -r ref_fofn
TE_detective nadiscover -i test_sim.bam -r ref_fofn --polyA --discord_cluster_dens 5
TE_detective analyze -i test_sim.bam -r ref_fofn --inp initial_predictions.txt
TE_detective cluster2d -i test_sim.bam -r ref_fofn
TE_detective filter -i final_results.tsv --bed rmsk_ucsc_mm10.bed
You can compare results to the files in TEdetective_example/example_results
This uses data from one of the CEU-trios to identify TE insertions found in the child, but not in either parent.
-
In your working directory ($DIR) you need four subdirectories: one for each sample, and then one for polymorphic analysis (NA12878, NA12891, NA12892, polymorph)
-
You have downloaded, aligned and sorted the bam file for each of the three individuals using the instructions above. Referred to as NA12878_hg19_sorted.bam, NA12891_hg19_sorted.bam, and NA12892_hg19_sorted.bam below.
-
Create a file called ref_fofn in $DIR that has a single line with the repeat element and location (complete path) of a fasta file for the repeat element of interest. See above and/or example ref_fofn file in example_data.
-
obtain a bed formatted file of annotated TE entries. Called rmsk_hg19.bed below. See info above.
mkdir -p $DIR/NA12878
mkdir -p $DIR/NA12891
mkdir -p $DIR/NA12892
mkdir -p $DIR/polymorph
cp $PATH_TO_FILE/ref_fofn $DIR
cp $PATH_TO_FILE/rmsk_hg19.bed $DIR
cd $DIR/NA12878/
TE_detective preprocess -i $PATH_TO_FILE/NA12878_hg19_sorted.bam -r ../ref_fofn
TE_detective discover -i $PATH_TO_FILE/NA12878_hg19_sorted.bam -r ../ref_fofn --read_length 100 --insert_size_est 383 --coverage_cutoff 608
TE_detective nadiscover -i $PATH_TO_FILE/NA12878_hg19_sorted.bam -r ../ref_fofn -o initial_predictions_NA12878.txt --polyA --read_length 100 --insert_size_est 383 --discord_cluster_dens 10 --coverage_cutoff 608
TE_detective analyze -i $PATH_TO_FILE/NA12878_hg19_sorted.bam -r ../ref_fofn -o final_results_NA12878.txt --inp initial_predictions_NA12878.txt --read_length 100 --insert_size_est 383
TE_detective filter -i final_results_NA12878.txt -b ../rmsk_hg19.bed
cd $DIR/NA12891/
TE_detective preprocess -i $PATH_TO_FILE/NA12891_hg19_sorted.bam -r ../ref_fofn
TE_detective discover -i $PATH_TO_FILE/NA12891_hg19_sorted.bam -r ../ref_fofn --read_length 100 --insert_size_est 439 --coverage_cutoff 504
TE_detective nadiscover -i $PATH_TO_FILE/NA12891_hg19_sorted.bam -r ../ref_fofn -o initial_predictions_NA12891.txt --polyA --read_length 100 --insert_size_est 439 --discord_cluster_dens 10 --coverage_cutoff 504
TE_detective analyze -i $PATH_TO_FILE/NA12891_hg19_sorted.bam -r ../ref_fofn -o final_results_NA12891.txt --inp initial_predictions_NA12878.txt --read_length 100 --insert_size_est 439
TE_detective filter -i final_results_NA12891.txt -b ../rmsk_hg19.bed
cd $DIR/NA12892/
TE_detective preprocess -i $PATH_TO_FILE/NA12892_hg19_sorted.bam -r ../ref_fofn
TE_detective discover -i $PATH_TO_FILE/NA12892_hg19_sorted.bam -r ../ref_fofn --read_length 100 --insert_size_est 439 --coverage_cutoff 504
TE_detective nadiscover -i $PATH_TO_FILE/NA12892_hg19_sorted.bam -r ../ref_fofn -o initial_predictions_NA12892.txt --polyA --read_length 100 --insert_size_est 439 --discord_cluster_dens 10 --coverage_cutoff 504
TE_detective analyze -i $PATH_TO_FILE/NA12892_hg19_sorted.bam -r ../ref_fofn -o final_results_NA12892.txt --inp initial_predictions_NA12878.txt --read_length 100 --insert_size_est 439
TE_detective filter -i final_results_NA12892.txt -b ../rmsk_hg19.bed
We use the bam and preprocessed files from the parents to analyze the initial predictions from the child. Then we apply the ceu filter sets as well as filter for annotated TEs.
cd $DIR/polymorph
TE_detective analyze -i $PATH_TO_FILE/NA12891_hg19_sorted.bam -r ../ref_fofn -o final_results_NA12878_NA12891.txt --inp ../NA12878/initial_predictions_NA12878.txt --read_length 100 --insert_size_est 439 -p ../NA12891/preprocessed_files --log_file analyze.91.log
TE_detective analyze -i $PATH_TO_FILE/NA12892_hg19_sorted.bam -r ../ref_fofn -o final_results_NA12878_NA12892.txt --inp ../NA12878/initial_predictions_NA12878.txt --read_length 100 --insert_size_est 439 -p ../NA12892/preprocessed_files --log_file analyze.92.log
TE_detective filter -i ../NA12878/final_results_NA12878.txt -s final_results_NA12878_NA12891.txt,final_results_NA12878_NA12892.txt --bed_screen ../rmsk_hg19.bed --filter ceu --pm_qual_thresh 0.8 --te_type LINE -o FINAL_RESULTS.PM.txt
We predict final TE insertions in the child and parents using the ceu filter set as well as filter for annotated TEs. We then filter all TEs from the child that are within insert_size_est of a TE predicted in either parent.
TE_detective filter -i ../NA12878/final_results_NA12878.txt --bed_screen ../rmsk_hg19.bed --filter ceu --pm_qual_thresh 0.8 --te_type LINE -o FINAL_RESULTS.NA12878.txt
TE_detective filter -i ../NA12891/final_results_NA12891.txt --bed_screen ../rmsk_hg19.bed --filter ceu --pm_qual_thresh 0.8 --te_type LINE -o FINAL_RESULTS.NA12891.txt
TE_detective filter -i ../NA12892/final_results_NA12892.txt --bed_screen ../rmsk_hg19.bed --filter ceu --pm_qual_thresh 0.8 --te_type LINE -o FINAL_RESULTS.NA12892.txt
TE_detective filter -i ../NA12878/final_results_NA12878.txt --results_screen_files FINAL_RESULTS.NA12891.txt,FINAL_RESULTS.NA12892.txt --bed_screen ../rmsk_hg19.bed --filter ceu --pm_qual_thresh 0.8 --te_type LINE -o FINAL_RESULTS.NORM.txt --insert_size_est 383
The final results from insertion prediction using polymorphic subtraction are in: FINAL_RESULTS.PM.txt
The final results from insertion prediction using normal subtraction are in: FINAL_RESULTS.NORM.txt
Censor is distributed under the GPL license. See details in Kohany et. al. Bioinformatics 2006.
NCBI Blast is freely available to the public for use as a "United States Government Work". See details here.
1. Preprocess
usage: TE_detective preprocess [-h] -i BAM_INP -r FOFN_REF [-p PREPROCESS_DIR]
[--min_clipped_len CLL_INP]
[--log_file LOG_FILE]
Processes the input files (indexed BAM file and indexed fasta file), extracts
discordant and clipped reads, as well as creates other files needed in
subsequent steps. Outputs all files to directory specified by --preprocess_dir
required arguments:
-i BAM_INP, --input_bam BAM_INP
input Bam(.bam) file of aligned reads
-r FOFN_REF, --ref FOFN_REF
File with reference sequence paths, see README.md for
more info
optional arguments:
-h, --help show this help message and exit
-p PREPROCESS_DIR, --preprocess_dir PREPROCESS_DIR
directory to store preprocessing output files
(default: preprocessed_files)
--min_clipped_len CLL_INP
Minimum clipped length(bp) (default: 25)
--log_file LOG_FILE run log file (default: preprocess.log)
2. Discover
usage: TE_detective discover [-h] -i BAM_INP -r FOFN_REF [-o OUTPUT_FILE]
[-p PREPROCESS_DIR] [--insert_size_est ISZ_INP]
[--read_length RDL_INP]
[--discord_cluster_dens DRD_INP]
[--coverage_cutoff CCT_INP]
[--min_clipped_len CLL_INP]
[--min_map_qual MPQ_INP]
[--map_qual_uniq MPQU_INP] [--log_file LOG_FILE]
Uses output from preprocessing step and makes an initial list of candidate
insertions
required arguments:
-i BAM_INP, --input_bam BAM_INP
Input Bam(.bam) file of aligned reads
-r FOFN_REF, --ref FOFN_REF
File with reference sequence paths, see README.md for
more info
optional arguments:
-h, --help show this help message and exit
-o OUTPUT_FILE, --output_file OUTPUT_FILE
Tab-delimited file of initial set of TE insertions
(default: initial_predictions.txt)
-p PREPROCESS_DIR, --preprocess_dir PREPROCESS_DIR
directory used to store preprocessing output files
(default: preprocessed_files)
--insert_size_est ISZ_INP
Insert size estimate (default: 340)
--read_length RDL_INP
Average read length (default: 150)
--discord_cluster_dens DRD_INP
Discord read cluster density (default: 10)
--coverage_cutoff CCT_INP
Coverage cutoff input (default: 200)
--min_clipped_len CLL_INP
Minimum clipped length(bp) (default: 25)
--min_map_qual MPQ_INP
Minimum mapping quality (default: 30)
--map_qual_uniq MPQU_INP
Minimum mapping quality (default: 1)
--log_file LOG_FILE run log file (default: discover.log)
3. Nadiscover
usage: TE_detective nadiscover [-h] -i BAM_INP -r FOFN_REF [--bed RMSK_BED]
[-o OUTPUT_FILE] [-p PREPROCESS_DIR]
[--min_clipped_len CLL_INP]
[--insert_size_est ISZ_INP]
[--read_length RDL_INP]
[--discord_cluster_dens DRD_INP]
[--coverage_cutoff CCT_INP] [--all]
[--merge_aligned] [--nonaligned_search]
[--min_map_qual MPQ_INP]
[--map_qual_uniq MPQU_INP] [--polyA]
[--polyA_len PQL_INP]
[--polyA_mismatch PMM_INP]
[--log_file LOG_FILE]
Performs nonalignment part of the discovery step. Module adds poly A/T
information into predictions made by discovery step. This module performs
initial searches as well, but without using BWA aligner for clipped and
discordant read alignment to TE reference sequence. Instead, a bed file of
masked regions is provided as input, and alignment information from input BAM
file is used.
required arguments:
-i BAM_INP, --input_bam BAM_INP
input Bam(.bam) file of aligned reads
-r FOFN_REF, --ref FOFN_REF
File with reference sequence paths, see README.md for
more info
optional arguments:
-h, --help show this help message and exit
--bed RMSK_BED FoFn for existing repeat elements
-o OUTPUT_FILE, --output_file OUTPUT_FILE
Tab-delimited output file of initial set of TE
insertions (default: initial_predictions_noalign.txt)
-p PREPROCESS_DIR, --preprocess_dir PREPROCESS_DIR
directory used to store preprocessing output files
(default: preprocessed_files)
--min_clipped_len CLL_INP
Minimum clipped length(bp) (default: 25)
--insert_size_est ISZ_INP
insert Size estimate (default: 340)
--read_length RDL_INP
Average read length (default: 150)
--discord_cluster_dens DRD_INP
discord read cluster density (default: 5)
--coverage_cutoff CCT_INP
Coverage cutoff input (default: 200)
--all use all reads instead of only clipped (default: False)
--merge_aligned Merge aligned predictions (default: False)
--nonaligned_search Perform non-alignment ref bam search (default: False)
--min_map_qual MPQ_INP
Minimum mapping quality (default: 30)
--map_qual_uniq MPQU_INP
Minimum mapping quality unique test (default: 1)
--polyA Perform poly A/T search (default: False)
--polyA_len PQL_INP poly A/T Length (default: 9)
--polyA_mismatch PMM_INP
poly A/T mismatch (default: 1)
--log_file LOG_FILE run log file (default: nadiscover.log)
4. Analyze (Realignment step from figure 2)
usage: TE_detective analyze [-h] -i BAM_INP -r FOFN_REF --inp LIST_INP
[-p PREPROCESS_DIR] [-o OUTPUT_FILE]
[--read_length RDL_INP]
[--min_clipped_len CLL_INP]
[--min_anchor_len AHL_INP]
[--clipped_read_range CER_INP]
[--clipped_search_interval CSI_INP]
[--min_breakpt_reads MRE_INP]
[--min_het_reads MRH_INP]
[--insert_size_est ISZ_INP]
[--mapping_qual_interval QII_INP]
[--intervals NII_INP] [--min_map_qual MPQ_INP]
[--map_qual_uniq MPQU_INP]
[--filter_discord_mates] [--log_file LOG_FILE]
Realigns reads around a predicted insertion point. Can be used to refine
initial predictions from the discover step, or to find evidence of potential
insertions in a different sample (e.g. for polymorphic subtraction). Filter
output from detailed analysis section. User can filter results using the
filter module or manually by importing them into Excel or any other tool
required arguments:
-i BAM_INP, --input_bam BAM_INP
input Bam(.bam) file of aligned reads
-r FOFN_REF, --ref FOFN_REF
File with reference sequence paths, see README.md for
more info
--inp LIST_INP Input list of insertions
optional arguments:
-h, --help show this help message and exit
-p PREPROCESS_DIR, --preprocess_dir PREPROCESS_DIR
directory used to store preprocessing output files
(default: preprocessed_files)
-o OUTPUT_FILE, --output_file OUTPUT_FILE
Tab-delimited output file of potential TE
insertions(default: final_resutls.tsv)
--read_length RDL_INP
Average read length (default: 150)
--min_clipped_len CLL_INP
Minimum clipped length(bp) (default: 25)
--min_anchor_len AHL_INP
Minimum anchor length(bp) (defualt: 30)
--clipped_read_range CER_INP
Range of clipped reads at a end to put in a group
(default: 5)
--clipped_search_interval CSI_INP
Clipped read search interval (default: 20)
--min_breakpt_reads MRE_INP
Min read for breakpoint (default: 4)
--min_het_reads MRH_INP
Minimum reads to call hetrozygous insertion (default:
3)
--insert_size_est ISZ_INP
Insert size estimate (default: 340)
--mapping_qual_interval QII_INP
Interval for mapping quality (default: 0.05)
--intervals NII_INP Number of intervals (default: 6)
--min_map_qual MPQ_INP
Minimum mapping quality (default: 30)
--map_qual_uniq MPQU_INP
Minimum mapping quality uniq test (default: 1)
--filter_discord_mates
Filter discord mate files (default: False)
--log_file LOG_FILE run log file (default: analyze.log)
5. Cluster2D
usage: TE_detective cluster2d [-h] -i BAM_INP -r FOFN_REF [-o OUTPUT_FILE]
[-p PREPROCESS_DIR] [--insert_size_est ISZ_INP]
[--read_length RDL_INP]
[--discord_cluster_dens DRD_INP]
[--coverage_cutoff CCT_INP] [--all]
[--log_file LOG_FILE]
Optional module to change the discordant read clustering density for initial
prediction without realigning everything. For example, if
--discord_cluster_dens was set to 10 for initial discovery step and user want
to see predictions with --discord_cluster_dens = 5. Uses intermediate files
from discover section and generates new prediction file.
required arguments:
-i BAM_INP, --input_bam BAM_INP
input Bam(.bam) file of aligned reads
-r FOFN_REF, --ref FOFN_REF
File with reference sequence paths, see README.md for
more info
optional arguments:
-h, --help show this help message and exit
-o OUTPUT_FILE, --output_file OUTPUT_FILE
Tab-delimited file of initial set of TE insertions
(default: recluster_initial_predictions.txt)
-p PREPROCESS_DIR, --preprocess_dir PREPROCESS_DIR
directory used to store preprocessing output files
(default: preprocessed_files)
--insert_size_est ISZ_INP
insert Size estimate (default: 340)
--read_length RDL_INP
Average read length (default: 150)
--discord_cluster_dens DRD_INP
Discord read cluster density (default: 5)
--coverage_cutoff CCT_INP
Coverage cutoff input (default: 200)
--all use all reads instead of only clipped (default: False)
--log_file LOG_FILE run log file (default: cluster2d.log)
6. Filter
usage: TE_detective filter [-h] -i OFA_INP -b FOFN_BED [-p PREPROCESS_DIR]
[--align_qual_lim QLM_INP]
[--min_clipped_reads TCR_INP]
[--min_clipped_and_dischord_reads TRD_INP]
[--read_percent RP_INP] [--read_length RDL_INP]
[--insert_size_est ISZ_INP] [--log_file LOG_FILE]
Filter output from analyze step.# Filteration step code looks like this if
total_clipped_rd >= tcr or ( (total_clipped_rd >= mtcr ) and (
(total_clipped_rd_wpat+total_discord_rd) >= trd ) ): filter_result = 'PASS'
elif total_discord_rd >= odrd: filter_result = 'PASS_D' # This flag says
passed based on only discordant reads.
required arguments:
-i OFA_INP, --input_file OFA_INP
use the output file from analyze section as input
-b FOFN_BED, --bed FOFN_BED
File containg a list of files to existing repeat
elements. List the full path for each file. See
example in example_data
optional arguments:
-h, --help show this help message and exit
-p PREPROCESS_DIR, --preprocess_dir PREPROCESS_DIR
directory used to store preprocessing output files
(default: preprocessed_files)
--align_qual_lim QLM_INP
Lowest limit for alignment quality (default: 0.85)
--min_clipped_reads TCR_INP
Minimum number of clipped reads (default: 5)
--min_clipped_and_dischord_reads TRD_INP
Minimum total [clipped+discordant] reads (default: 10)
--read_percent RP_INP
read percent value (default: 10.0)
--read_length RDL_INP
Average read length (default: 150)
--insert_size_est ISZ_INP
insert Size estimate (default: 340)
--log_file LOG_FILE run log file (default: filter.log)