This protocol computes Whole Genome Alignments (WGA) to discover syntenic SNPs out of reads mapped to concatenated genome references. It requires FASTA and VCF input files and produces multiple sequence alignments of subgenomes that make up polyploids.
Rubén Sancho (1,2), Pilar Catalán (2), Bruno Contreras Moreira (1,3)
- Estación Experimental de Aula Dei-CSIC, Zaragoza, Spain
- Escuela Politécnica Superior de Huesca, U.Zaragoza, Spain
- Fundación ARAID, Zaragoza, Spain
While this protocol was developed on Linux x86_64 systems, it should also work on MacOS settings. It requires some standard Linux utilities (gzip, sort, perl, make, python3, g++, etc) and a few third-party dependencies which can be installed locally as follows:
git clone https://github.com/eead-csic-compbio/vcf2alignment.git
cd vcf2alignment
make install
# optionally, takes a couple minutes
make test
The table shows the main dependencies of this package:
| software | flag | source | notes |
|---|---|---|---|
| Cgaln | https://github.com/rnakato/Cgaln | requires gcc compiler | |
| GSAlign | -g | https://github.com/hsinnan75/GSAlign | requires g++ compiler |
| Red | skipped with -m | https://github.com/EnsemblGenomes/Red | requires g++ compiler |
| Red2Ensembl.py | https://github.com/Ensembl/plant-scripts | requires python3 | |
| gnuplot | http://www.gnuplot.info | required for dotplots in PDF format |
Other Linux dependencies required include: wget python3 g++ gnuplot-qt libdb-dev
See ci.yml.
These are the data files required to run this pipeline:
-
2+ FASTA files of genome assemblies of diploid species of the taxa of interest, one file per species. Our Brachypodium sample data includes the GZIP compressed genomes of Brachypodium distachyon and Brachypodium stacei, which are named
Bdis.fna.gzandBsta.fna.gz. Each file should have unique chromosome names. -
A VCF file with GBS/RADseq/RNAseq/WGS sequence reads of presumably polyploids mapped to the concatenated previously described FASTA files. See file
BdisBd2_BstaChr01.vcf.gzin sample data for an example.
A typical analysis involves calling three scripts: WGA, vcf2alignment and vcf2synteny.
Note that the steps below can be run in one go as follows:
make test
WGAs must be computed to find syntenic segments among the reference genomes available for read mapping. By default this uses CGaln, which requires the input sequences to be soft-masked ahead. In our example we set B. distachyon as the master reference genome for being the best quality assembly at hand (see 2.3 below). In this we now find out syntenic segments on the other genomes, defined as secondary genomes (B. stacei). Each individual reference is hence considered a subgenome to which reads map:
./WGA -A sample_data/Bdis.fna.gz -B sample_data/Bsta.fna.gz
This produces the following output:
## WGA -A sample_data/Bdis.fna.gz -B sample_data/Bsta.fna.gz -l 1 -m 1 -G 0 -I -K11 -BS10000 -C -X12000 -fc -cons -n 4
## root: Bdis.fna.gz.Bsta.fna.gz_Cgaln_-K11_-BS10000_-X12000_-fc_-cons
# filter_FASTA_sequences: [passed 59130575 bp] Bd2
# filter_FASTA_sequences: [passed 31564145 bp] Chr01
# chrcode A example: Bd(\d+)
# chrcode B example: Chr(\d+)
## soft-masking filtered sequences
# filtered file A: sample_data/Bdis.fna.gz.sm.fasta
# filtered file B: sample_data/Bsta.fna.gz.sm.fasta
## CGaln algorithm
## indexing masked, filtered sequences
[...]
## computing and plotting Whole Genome Alignment
[...]
## converting Whole Genome Alignment to BED
## output files:
# BED: Bdis.fna.gz.Bsta.fna.gz_Cgaln_-K11_-BS10000_-X12000_-fc_-cons.bed
# LOG: Bdis.fna.gz.Bsta.fna.gz_Cgaln_-K11_-BS10000_-X12000_-fc_-cons.coords.log
# PDF: Bdis.fna.gz.Bsta.fna.gz_Cgaln_-K11_-BS10000_-X12000_-fc_-cons.dot.pdf
## WGA summary: valid blocks: 54 unique positions: 8956568
# time used (s): 119 memory used (Mb): 407.9
The most important result files are the 0-based BED list of syntenic positions, which will be used in the last step,
and the PDF dotplot, which requires gnuplot in your syste, which must be inspected to assess the quality of
the WGA. Note that flags -I and -C can be used to tweak the WGA parameters after inspection of the dotplot.
Alternatively, the GSAlign WGA algorithm can be invoked as follows, adding optionally flag -G to adapt the default parameters to your genomes of interest:
./WGA -A sample_data/Bdis.fna.gz -B sample_data/Bsta.fna.gz -g
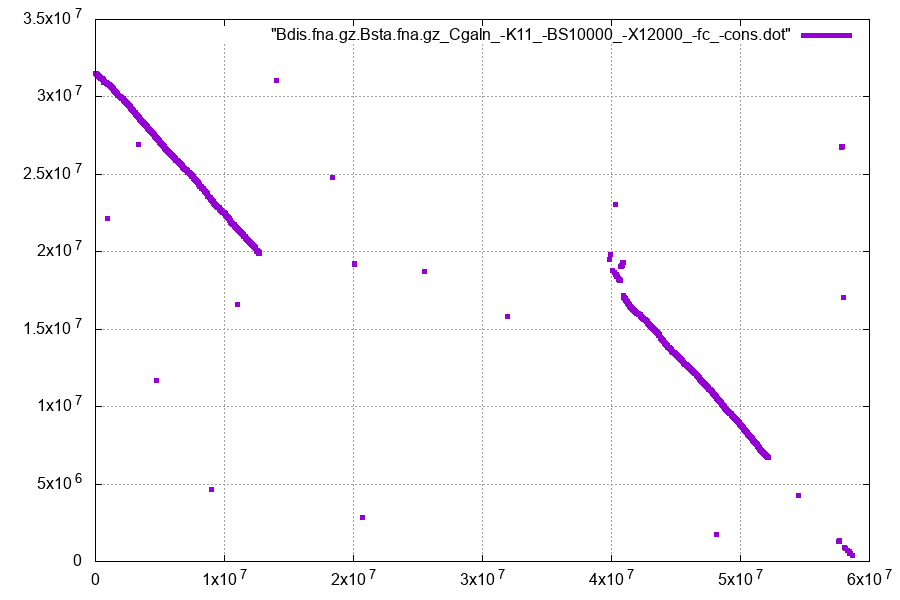 Figure 1. WGA dotplot resulting from Cgaln alignment of two homologous chromosomes.
Figure 1. WGA dotplot resulting from Cgaln alignment of two homologous chromosomes.
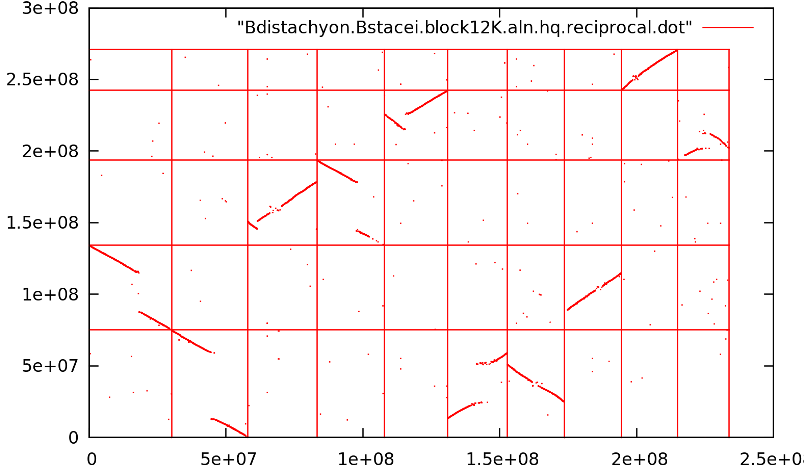 Figure 2. Multi-chromosome WGA dotplot. A good WGA dotplot will have long diagonal runs of aligned genome regions instead of clouds.
Figure 2. Multi-chromosome WGA dotplot. A good WGA dotplot will have long diagonal runs of aligned genome regions instead of clouds.
Script vcf2alignment parses an input VCF file, with might be GZIP/BZIP2 compressed,
and produces a list of valid sites considering min read depth (-d) and max missing data (-m).
Note this requires a config file that matches sample names in the VCF file to their human-readable names
(see example sample_data/config.tsv).
A report log file with valid 1-based coordinates (-l) is saved to be used in the last step:
./vcf2alignment -v sample_data/BdisBd2_BstaChr01.vcf.gz -c sample_data/config.tsv -l BdisBd2_BstaChr01.vcf.log -d 5 -m 3
Script vcf2synteny puts it all together and produces an alignment in FASTA format:
./vcf2synteny -v sample_data/BdisBd2_BstaChr01.vcf.gz -c sample_data/config.synteny.tsv -l BdisBd2_BstaChr01.vcf.log \
-d 5 -m 3 -r Bdis -o BdisBd2_BstaChr01.DP5.M3.synteny.fasta
Note that a different config file is now used (see example sample_data/config.synteny.tsv), which also contains:
- a path to the BED file obtained in step 2.1
- regular expressions to match chromosome names from reference genomes used in step 2.1, can use those proposed by
WGA
This produces the following output:
# computing Bdis.Bsta.coords.positions.tsv (3 steps)
# master reference: Bdis
# secondary references: Bsta
# synteny files (SYNTENYZEROBASED=1):
# Bsta : Bdis.Bsta.coords.positions.tsv...
# total positions=780747
# decompressing VCF file with GZIP
# number of samples found=6
# number of loci read from VCF: 10000
...
# number of loci read from VCF: 780000
# sorting SNPs by position ...
# aligned position: Bd2_8991545 : Chr01_4671897,
# aligned position: Bd2_8991546 : Chr01_4671898,
...
# aligned position: Bd2_51018397 : Chr01_7785782,
# number of valid loci=781827
# number of polymorphic loci=9769
# Bdis_ABR2_Bdis variants: 1080 / 780747
# Bdis_ABR2_Bsta variants: 8991 / 780747
# Bdis_Bd21Control_Bdis variants: 1080 / 780747
# Bdis_Bd21Control_Bsta variants: 4048 / 780747
# Bhyb_Bhyb26_Bdis variants: 1076 / 780747
# Bhyb_Bhyb26_Bsta variants: 758944 / 780747
# Bhyb_ABR113_Bdis variants: 1010 / 780747
# Bhyb_ABR113_Bsta variants: 692978 / 780747
# Bsta_ABR114_Bdis variants: 17 / 780747
# Bsta_ABR114_Bsta variants: 745965 / 780747
# Bsta_TE4.3_Bdis variants: 0 / 780747
# Bsta_TE4.3_Bsta variants: 773119 / 780747
# concatenating temp files ...
# time used (s): 107 memory used (Mb): 658.1
The resulting multiple sequence alignment (MSA) has as many lines per sample as references, which are handled as subgenomes. The first 200 positions of the MSA derived from the sample data looks as follows:
>Bdis_ABR2_Bdis
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTCNNNNANNCNNNNNNNNNNNNNNNCNTNNNNTNNNNNNNNNNNNNGNTNNNNNTNNGGCNNNNNNNNNNNNNNGANANCC
>Bdis_ABR2_Bsta
ATCTCGCGGCTGCCCCCCCGAGTTCGGAGGACCACCGGCCTCGCCGGGCCTCCACGTCGTGGCCACTGCTTCGGCAACGCACTGCTGACCTCACCGCTGCCGTCGCACTGGCAAACGGGTCAGCAAATCAAGCGCGCTGCGTGTCGTCNCGTCTCGGGCCATGCCGCTTTTCATCTGGCCGCCCTGGTTGCGCGACACCC
>Bdis_Bd21Control_Bdis
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTCNNNNANNCNNNNNNNNNNNNNNNCNTNNNNTNNNNNNNNNNNNNGNTNNNNNTNNGGCNNNNNNNNNNNNNNGANANCC
>Bdis_Bd21Control_Bsta
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
>Bhyb_Bhyb26_Bdis
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTCNNNNANNCNNNNNNNNNNNNNNNCNTNNNNTNNNNNNNNNNNNNGNTNNNNNTNNGGCNNNNNNNNNNNNNNGANANCC
>Bhyb_Bhyb26_Bsta
ATCTCGCGGCTGCCCCCCCGAGTTCGGAGGACCACCGGCCTCGCCGGGCCTCCACGTCGTGGCCACTGCTTCGGCAACGCACTGCTGACCTCACCGCTGCCGTCGCACTGGCAAACGGGTCAGCAAATCAAGCGCGCTGCGTGTCNTCGCGTCTCGGGCCATGCCGCTTTTCATCTGGCCGCCCTGGTTGCGCGACACCC
>Bhyb_ABR113_Bdis
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
>Bhyb_ABR113_Bsta
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
>Bsta_ABR114_Bdis
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGNTNNNNNTNNGGCNNNNNNNNNNNNNNGANANCC
>Bsta_ABR114_Bsta
ATCTCGCGGCTGCCCCCCCGAGTTCGGAGGACCACCGGCCTCGCCGGGCCTCCACGTCGTGGCCACTGCTTCGGCAACGCACTGCTGACCTCACCGCTGCCGTCGCACTGGCAAACGGGTCAGCAAATCAAGCGCGCTGCGTGTCGTCGCGTCTCGGGCCATGCCGCTTTTCATCTGGCCGCCCTGGTTGCGCGACACCC
>Bsta_TE4.3_Bdis
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
>Bsta_TE4.3_Bsta
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAATCAAGCGCGCTGCGTGTCGTCGCGTCTCGGGCCATGCCGCTTTTCATCTGGCCGCCCTGGTTGCGCGACACCC
this paper, to be written
Lin HN, Hsu WL (2020) GSAlign: an efficient sequence alignment tool for intra-species genomes. BMC Genomics 21:182. https://doi.org/10.1186/s12864-020-6569-1
Nakato R, Gotoh O (2010) Cgaln: fast and space-efficient whole-genome alignment. BMC Bioinformatics 11:224. https://doi.org/10.1186/1471-2105-11-224
Girgis HZ (2015) Red: an intelligent, rapid, accurate tool for detecting repeats de-novo on the genomic scale. BMC Bioinformatics 16:227. https://doi.org/10.1186/s12859-015-0654-5
Contreras-Moreira B, Filippi CV, Naamati G, García Girón C, Allen JE, Flicek P (2021) Efficient masking of plant genomes by combining kmer counting and curated repeats Genomics. Plant Genome https://doi.org/10.1002/tpg2.20143