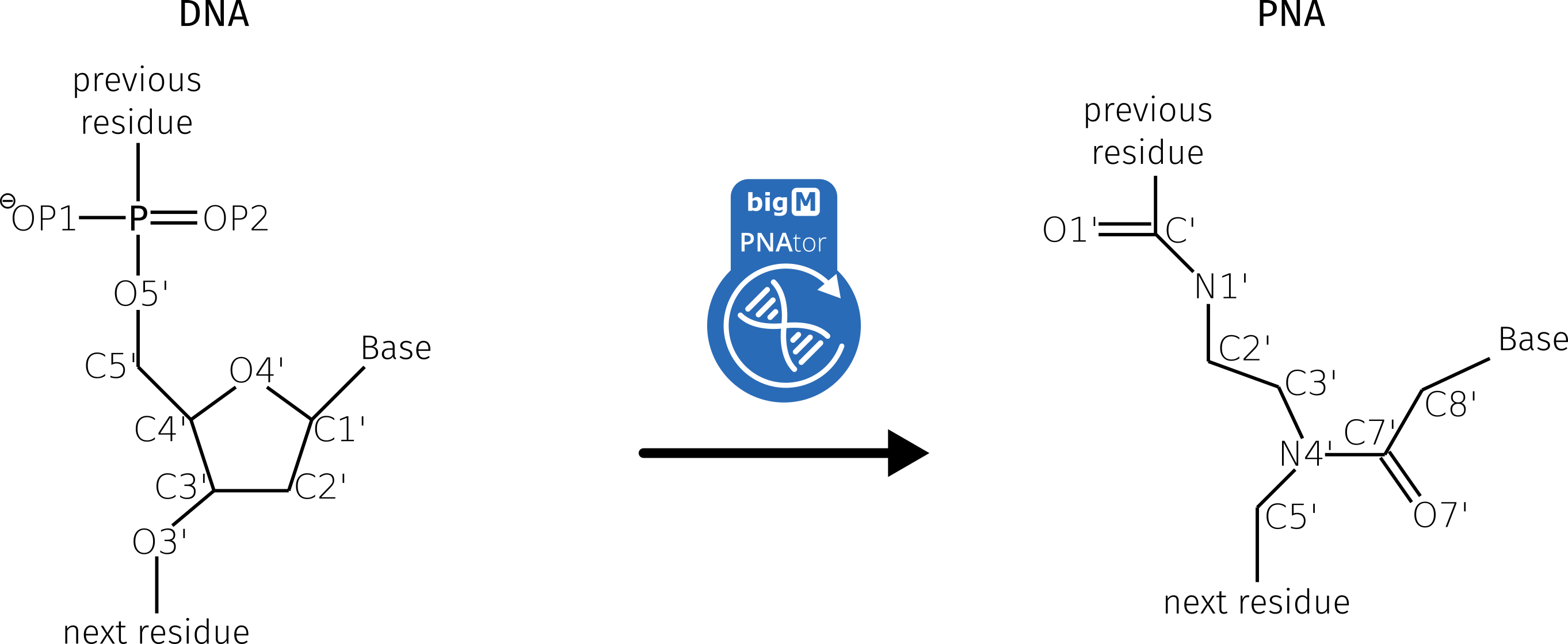
Peptide nucleic acid (PNA) are artificial DNAs or RNAs with an uncharged pseudopeptide backbone. In 1991, the PNA was described for the first time by Nielsen et al. (1). The sugar phosphate backbone is completely replaced by the polyamide backbone. The backbone of the PNA consists of 2-(aminoethyl)glycine units, the nucleobases adenine, cytosine, guanine, thymine and uracil are covalently linked to the backbone by an ethenone (1).
In 1998, Sen and Nielsen published a theoretical approach describing how DNA can be converted into PNA (2). This approach was based on four main steps: I) exchanging atoms of the backbone, II) removing atoms that are not present in the PNA structure, III) calculating missing atoms and VI) adding missing hydrogen atoms. The last step (nr. VI) was not implemented. Hydrogen atoms can be added with Ambertools16 (3).
For the implementation the SiNGA Framework was used to develop the PNAtor. The SiNGA Framework allows a comfortable handling by predefined methods and functions for the handling of 3D structures.
RNA structures generated with the RNAComposer (4) can also be converted into PNA.
References:
(1) Nielsen, Peter E., et al. "Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide." Science 254.5037 (1991): 1497-1500.
(2) Sen, Srikanta, and Lennart Nilsson. "Molecular dynamics of duplex systems involving PNA: structural and dynamical consequences of the nucleic acid backbone." Journal of the American Chemical Society 120.4 (1998): 619-631.
(3) Case, D. A., et al. "AmberTools 16, University of California, San Francisco, 2016." There is no corresponding record for this reference.
(4) Popenda, Mariusz, et al. "Automated 3D structure composition for large RNAs." Nucleic acids research 40.14 (2012): e112-e112.
- clone repro
- navigate to PNAGenerator.java in IDE of your choice
- choose your PDB Structure (DNA/RNA)
Structure structure = StructureParser.online()
.pdbIdentifier("1OLD")
.everything()
.setOptions(StructureParserOptions.withSettings(
StructureParserOptions.Setting.OMIT_HYDROGENS))
.parse();- execute the
convertToPNAStructure(structure)method - write reulst file
try {
StructureWriter.writeLeafSubstructureContainer(structure.getFirstModel(),
Paths.get("path/to/outputPDB.pdb"));
} catch (IOException e) {
e.printStackTrace();
}Make sure you have the Java 8 or later installed.