A new anomaly detection algorithm that brings together the best from ANODE and CWoLa. Train a density estimator on sidebands, sample artificial datapoints in the signal region, train a classifier to distinguish artificial and real signal region data and then use the same classifier for classifying signal (the anomaly) from background.
To see the definition of signal region and sideband region please see: SB-SR
Follow the instructions below to reproduce the results and/or perform further studies. The steps "Train the ANODE model", "Mix data and samples", "Train the classifier", and "Evaluation" can be called separately as described below. Alternatively, the script run_all.py can be used to run the full pipeline in one call.
If you use CATHODE for your research, please cite:
- "Classifying Anomalies THrough Outer Density Estimation (CATHODE)",
By Anna Hallin, Joshua Isaacson, Gregor Kasieczka, Claudius Krause, Benjamin Nachman, Tobias Quadfasel, Matthias Schlaffer, David Shih, and Manuel Sommerhalder.
arXiv:2109.00546.
(can be skipped if one starts directly from the preprocessed samples here)
To get the datasets:
wget https://zenodo.org/record/4536377/files/events_anomalydetection_v2.features.h5
wget https://zenodo.org/record/5759087/files/events_anomalydetection_qcd_extra_inneronly_features.h5
To preprocess:
python run_data_preparation_LHCORD.py
To scan over different signal injections and/or different splits, use the --S_over_B and --seed option respectively. The results in the paper when scanning into lower S/B ratios were achieved by varying the seed from 1 to 10.
Use the script run_all.py to run the full pipeline in one go. The flag --mode with the options CATHODE, ANODE, CWoLa, or supervised specifies which analysis type will be run. Explanation of additional arguments are explained when calling python run_all.py -h. In general, arguments considering the density estimator step start with --DE_, and arguments considering the classifier step start with --cf_.
The command to produce the most up-to-date performance is:
python run_all.py --data_dir separated_data/ --mode CATHODE --cf_separate_val_set --no_extra_signal --cf_n_samples 400000 --cf_realistic_conditional --cf_oversampling --cf_no_logit --cf_use_class_weights --cf_save_model --cf_n_runs 1
The corresponding script is run_ANODE_training.py
The corresponding script is run_classifier_data_creation.py
The corresponding script is run_classifier_training.py
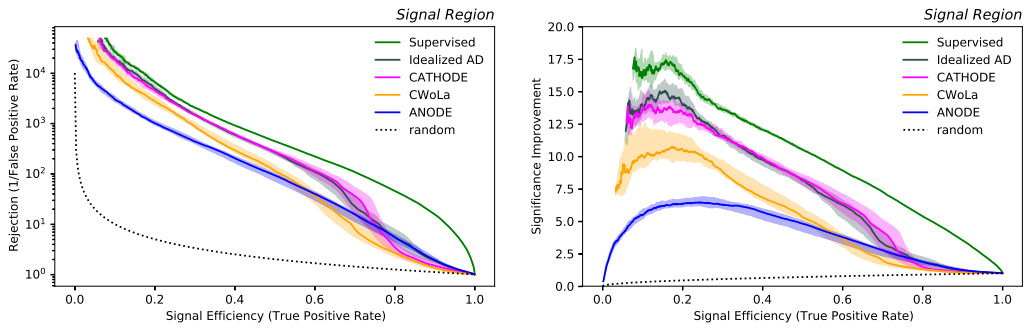
The evaluation leading to the main plots is shown in plotting_notebook.ipynb.
Alternatively, a short script like
from evaluation_utils import full_single_evaluation
data_savedir = 'classifier_data_folder/'
preds_dir = 'classifier_output_folder/'
_ = full_single_evaluation(data_savedir, preds_dir, n_ensemble_epochs=10, sic_range=(0, 20), savefig='result_SIC')
will plot the resulting SIC curve to file.
The most up-to-date command for the ANODE benchmark is:
python run_all.py --no_extra_signal --data_dir separated_data/ --mode ANODE
The most up-to-date command for the CWoLa Hunting benchmark is:
python run_all.py --data_dir separated_data/ --mode CWoLa --cf_separate_val_set --no_extra_signal --cf_oversampling --cf_no_logit --cf_use_class_weights --cf_save_model
The most up-to-date command for the idealized anomaly detector benchmark is:
python run_all.py --data_dir separated_data/ --mode idealized_AD --cf_separate_val_set --no_extra_signal --cf_no_logit --cf_oversampling --cf_use_class_weights --cf_save_model --cf_extra_bkg
The most up-to-date command for the fully supervised benchmark is:
python run_all.py --data_dir separated_data/ --mode supervised --cf_separate_val_set --no_extra_signal --cf_no_logit --cf_oversampling --cf_save_model --cf_extra_bkg
All the listed scripts provide documentation on how to use them by calling python [SCRIPT].py --help. In particular, the above example commands use default input/output directories and model names, which should be adjusted to custom choices when multiple studies are performed.