[Update August 2023 - data loading is now 10x faster!]
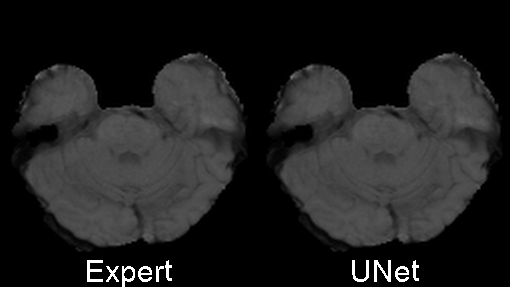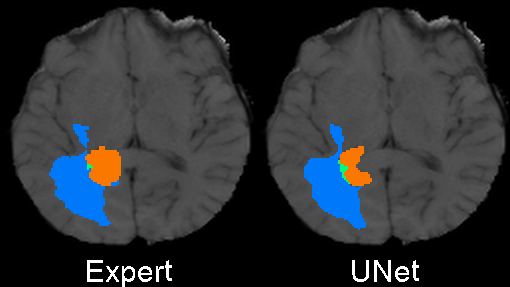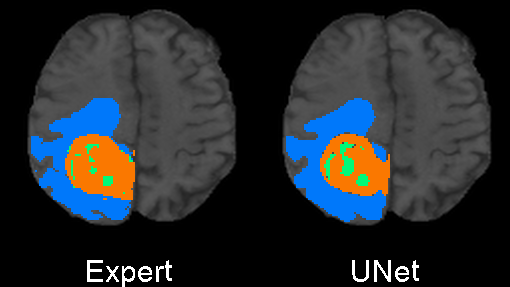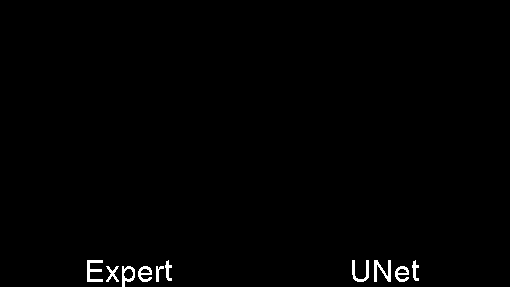
We designed 3DUnetCNN to make it easy to apply and control the training and application of various deep learning models to medical imaging data. The links above give examples/tutorials for how to use this project with data from various MICCAI challenges.
How to train a UNet on your own data.
-
Clone the repository:
git clone https://github.com/ellisdg/3DUnetCNN.git -
Install the required dependencies*:
pip install -r 3DUnetCNN/requirements.txt
*It is highly recommended that an Anaconda environment or a virtual environment is used to manage dependcies and avoid conflicts with existing packages.
See the Brats 2020 example for a description on how to create a configuration and train a model.
Once you have reviewed the documentation, feel free to raise an issue on GitHub, or email me at david.ellis@unmc.edu.
Ellis D.G., Aizenberg M.R. (2021) Trialing U-Net Training Modifications for Segmenting Gliomas Using Open Source Deep Learning Framework. In: Crimi A., Bakas S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2020. Lecture Notes in Computer Science, vol 12659. Springer, Cham. https://doi.org/10.1007/978-3-030-72087-2_4
Ellis D.G., Aizenberg M.R. (2020) Deep Learning Using Augmentation via Registration: 1st Place Solution to the AutoImplant 2020 Challenge. In: Li J., Egger J. (eds) Towards the Automatization of Cranial Implant Design in Cranioplasty. AutoImplant 2020. Lecture Notes in Computer Science, vol 12439. Springer, Cham. https://doi.org/10.1007/978-3-030-64327-0_6
Ellis, D.G. and M.R. Aizenberg, Structural brain imaging predicts individual-level task activation maps using deep learning. bioRxiv, 2020: https://doi.org/10.1101/2020.10.05.306951